
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 19,471,328 – 19,471,422 |
| Length | 94 |
| Max. P | 0.870078 |

| Location | 19,471,328 – 19,471,422 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.34 |
| Mean single sequence MFE | -18.77 |
| Consensus MFE | -13.21 |
| Energy contribution | -13.60 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.532704 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
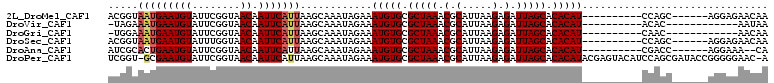
>2L_DroMel_CAF1 19471328 94 + 22407834 ACGGUAAUGAAUGUAUUCGGUAACAAUUCAUUAAGCAAAUAGAAAUGUGCGCUAAACGCAUUAAGAGAUUAGCACACAU----------CCAGC------AGGAGAACAA ....((((((((((........)).))))))))............((((.(((((.(.(.....).).))))).))))(----------((...------.)))...... ( -17.70) >DroVir_CAF1 32270 87 + 1 -UAGAAAUGAAUGUAUUCGGUAACAAUUCAUUAAGCAAAUAGAAAUGUGCGCUAAACGCAUUAAGAGAUUAGCACACAU----------ACAC------------AAUAA -....(((((((((........)).)))))))............(((((.(((((.(.(.....).).))))).)))))----------....------------..... ( -15.40) >DroGri_CAF1 29512 87 + 1 -UGGAAAUGAAUGUAUUCGGUAACAAUUCAUUAAGCAAAUAGAAAUGUGCGCUAAACGCAUUAAGAGAUUAGCACACAU----------CAAC------------AACAA -....(((((((((........)).)))))))............(((((.(((((.(.(.....).).))))).)))))----------....------------..... ( -14.80) >DroSec_CAF1 27030 94 + 1 ACGGUAAUGAAUGUAUUUGGUAACAAUUCAUUAAGCAAAUAGAAAUGUGCGCUAAACGCAUUAAGAGAUUAGCACACAU----------CCAGC------AGGAGAACAA ....((((((((((........)).))))))))............((((.(((((.(.(.....).).))))).))))(----------((...------.)))...... ( -17.70) >DroAna_CAF1 25680 92 + 1 AUCGCACUGAAUGUAUUCGGUAACAAUUCAUUAAGCAAAUAGAAAUGUGCGCUAAACGCAUUAAGAGAUUAGCACACAU----------CGACC------AGGAAA--CA .(((.((((((....)))))).......................(((((.(((((.(.(.....).).))))).)))))----------)))..------.(....--). ( -17.30) >DroPer_CAF1 34034 108 + 1 UCGGU-GCGAAUGUAUUCGGUAACAAUUCAUUAAGCAAAUAGAAAUGUGCGCUAAACGCAUUAAGAGAUUAGCACACAUACGAGUACAUCCAGCGAUACCGGGGGAAC-A (((((-(((.(((((((((.......((((((.....))).)))(((((.(((((.(.(.....).).))))).))))).)))))))))....)).))))))......-. ( -29.70) >consensus ACGGUAAUGAAUGUAUUCGGUAACAAUUCAUUAAGCAAAUAGAAAUGUGCGCUAAACGCAUUAAGAGAUUAGCACACAU__________CCACC______AGGAGAACAA .....(((((((((........)).)))))))............(((((.(((((.(.(.....).).))))).)))))............................... (-13.21 = -13.60 + 0.39)
| Location | 19,471,328 – 19,471,422 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.34 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -17.50 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.870078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
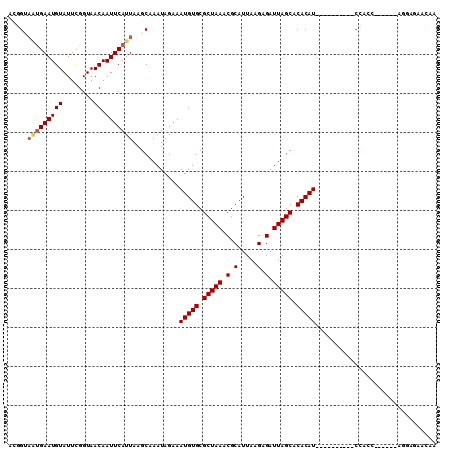
>2L_DroMel_CAF1 19471328 94 - 22407834 UUGUUCUCCU------GCUGG----------AUGUGUGCUAAUCUCUUAAUGCGUUUAGCGCACAUUUCUAUUUGCUUAAUGAAUUGUUACCGAAUACAUUCAUUACCGU ..........------...((----------(((((((((((.(.(.....).).)))))))))))...........((((((((.((........)))))))))))).. ( -22.20) >DroVir_CAF1 32270 87 - 1 UUAUU------------GUGU----------AUGUGUGCUAAUCUCUUAAUGCGUUUAGCGCACAUUUCUAUUUGCUUAAUGAAUUGUUACCGAAUACAUUCAUUUCUA- .....------------....----------(((((((((((.(.(.....).).)))))))))))............(((((((.((........)))))))))....- ( -18.30) >DroGri_CAF1 29512 87 - 1 UUGUU------------GUUG----------AUGUGUGCUAAUCUCUUAAUGCGUUUAGCGCACAUUUCUAUUUGCUUAAUGAAUUGUUACCGAAUACAUUCAUUUCCA- .....------------...(----------(((((((((((.(.(.....).).))))))))))))...........(((((((.((........)))))))))....- ( -19.60) >DroSec_CAF1 27030 94 - 1 UUGUUCUCCU------GCUGG----------AUGUGUGCUAAUCUCUUAAUGCGUUUAGCGCACAUUUCUAUUUGCUUAAUGAAUUGUUACCAAAUACAUUCAUUACCGU ..........------...((----------(((((((((((.(.(.....).).)))))))))))...........((((((((.((........)))))))))))).. ( -22.20) >DroAna_CAF1 25680 92 - 1 UG--UUUCCU------GGUCG----------AUGUGUGCUAAUCUCUUAAUGCGUUUAGCGCACAUUUCUAUUUGCUUAAUGAAUUGUUACCGAAUACAUUCAGUGCGAU ..--.....(------((..(----------(((((((((((.(.(.....).).)))))))))))).))).((((....(((((.((........)))))))..)))). ( -23.30) >DroPer_CAF1 34034 108 - 1 U-GUUCCCCCGGUAUCGCUGGAUGUACUCGUAUGUGUGCUAAUCUCUUAAUGCGUUUAGCGCACAUUUCUAUUUGCUUAAUGAAUUGUUACCGAAUACAUUCGC-ACCGA .-.......((((...((.(((((((.(((.(((((((((((.(.(.....).).)))))))))))(((.(((.....)))))).......))).)))))))))-)))). ( -29.90) >consensus UUGUUCUCCU______GCUGG__________AUGUGUGCUAAUCUCUUAAUGCGUUUAGCGCACAUUUCUAUUUGCUUAAUGAAUUGUUACCGAAUACAUUCAUUACCAU ...............................(((((((((((.(.(.....).).)))))))))))...........((((((((.((........)))))))))).... (-17.50 = -18.12 + 0.61)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 12:24:52 2006