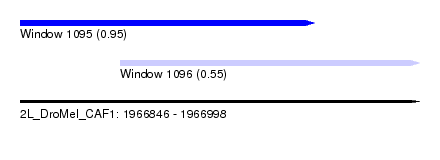
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,966,846 – 1,966,998 |
| Length | 152 |
| Max. P | 0.952710 |

| Location | 1,966,846 – 1,966,958 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.44 |
| Mean single sequence MFE | -27.57 |
| Consensus MFE | -18.04 |
| Energy contribution | -18.23 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
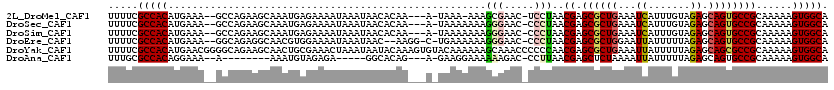
>2L_DroMel_CAF1 1966846 112 + 22407834 UUUUCGCCACAUGAAA--GCCAGAAGCAAAUGAGAAAAUAAAUAACACAA---A-UAAA-AAAGCGAAC-UCCUAACGAGCGCUGAAAUCAUUUGUAGAGCAGUGCCGCAAAAAGUGGCA .....(((((......--((.....)).......................---.-....-...(((...-..(....).((((((...((.......)).))))))))).....))))). ( -24.00) >DroSec_CAF1 9316 113 + 1 UUUUCGCCACAUGAAA--GCCAGAAGCAAAUGAGAAAAUAAAUAACACAA---A-UAAAAAAAGGGAAC-CCCUAACGAGCGCUGAAAUCAUUUGUAGAGCAGUGCCGCAAAAAGUGGCA .....(((((......--((.....)).......................---.-.......((((...-))))..((.((((((...((.......)).))))))))......))))). ( -25.20) >DroSim_CAF1 13467 113 + 1 UUUUCGCCACAUGAAA--GCCAGAAGCAAAUGAGAAAAUAAAUAACACAA---A-UAAAAAAAGGGAAC-CCCUAACGAGCGCUGAAAUCAUUUGUAGAGCAGUGCCGCAAAAAGUGGCA .....(((((......--((.....)).......................---.-.......((((...-))))..((.((((((...((.......)).))))))))......))))). ( -25.20) >DroEre_CAF1 18403 113 + 1 UUUUCGCCACAUGAAA--GGCAGAGGCAACGUGGAAAAUAAAUAAC--AAGG-C-UGAAAAAAGGGAAC-CCCUAACGAGCGCUGGAAUUAUUUUUAGAGCAGUGCCGCAAAAAGUGGCA .....(((((......--((((..(....)((.((((((((....(--(..(-(-(......((((...-))))....)))..))...))))))))...))..)))).......))))). ( -28.42) >DroYak_CAF1 16331 120 + 1 UUUUCGCCACAUGAACGGGGCAGAAGCAACUGCGAAACUAAAUAAUACAAAGUGUACAAAAAAGCAAACCCCCCAACGAGCGCUGAAAUUAUUUUUAGAGCAGCGCCGCAAAAAGUGGCA .....(((((......(((((((......))))............(((.....)))...............)))..((.((((((...(((....)))..))))))))......))))). ( -32.60) >DroAna_CAF1 40044 100 + 1 UUUGCGCCACAGGAAA--A--------AAAUGUAGAGA-----GGCACAG---A-GAAGGAAAAAAGAC-CCUUAACGAGCUCUAAAAUUAUUUUUAGAGCAGUGCCGCAAAAAGUGGCA .....(((((.(....--.--------...).....(.-----(((((.(---.-.((((.........-))))..)..(((((((((....))))))))).))))).).....))))). ( -30.00) >consensus UUUUCGCCACAUGAAA__GCCAGAAGCAAAUGAGAAAAUAAAUAACACAA___A_UAAAAAAAGGGAAC_CCCUAACGAGCGCUGAAAUCAUUUGUAGAGCAGUGCCGCAAAAAGUGGCA .....(((((.....................................................(((.....)))..((.((((((...((.......)).))))))))......))))). (-18.04 = -18.23 + 0.20)
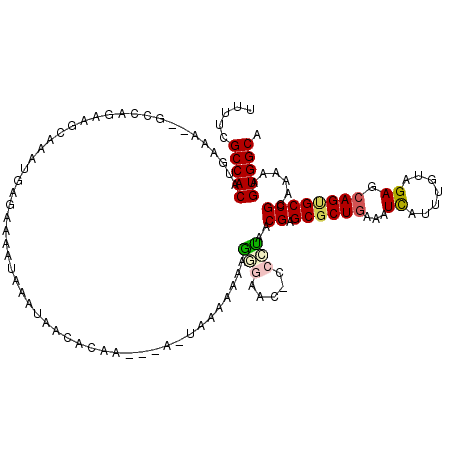

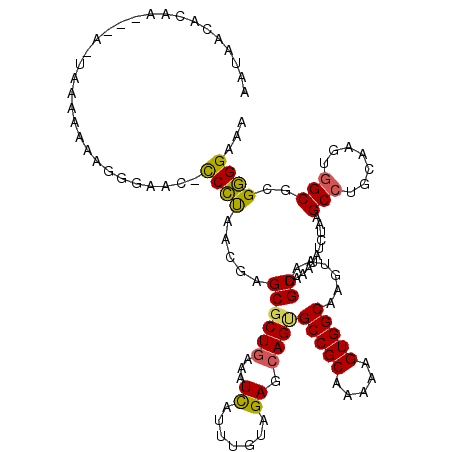
| Location | 1,966,884 – 1,966,998 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.80 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -21.88 |
| Energy contribution | -21.47 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.27 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.547846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
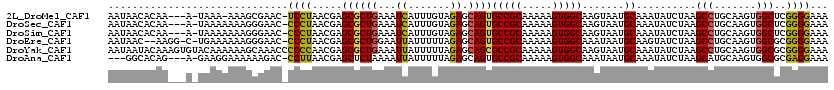
>2L_DroMel_CAF1 1966884 114 + 22407834 AAUAACACAA---A-UAAA-AAAGCGAAC-UCCUAACGAGCGCUGAAAUCAUUUGUAGAGCAGUGCCGCAAAAAGUGGCAAGUAAUGCAAAUAUCUAAGCCUGCAAGUGGCUCGGGGAAA ..........---.-....-.........-((((..(((((........(((((((((.((..((((((.....)))))).(.....)..........)))))))))))))))))))).. ( -31.00) >DroSec_CAF1 9354 115 + 1 AAUAACACAA---A-UAAAAAAAGGGAAC-CCCUAACGAGCGCUGAAAUCAUUUGUAGAGCAGUGCCGCAAAAAGUGGCAAGUAAUGCAAAUAUCUAAGCCUGCAAGUGGCUCGGGGAAA ..........---.-.............(-(((....((((........(((((((((.((..((((((.....)))))).(.....)..........)))))))))))))))))))... ( -31.60) >DroSim_CAF1 13505 115 + 1 AAUAACACAA---A-UAAAAAAAGGGAAC-CCCUAACGAGCGCUGAAAUCAUUUGUAGAGCAGUGCCGCAAAAAGUGGCAAGUAAUGCAAAUAUCUAAGCCUGCAAGUGGCUCGGGGAAA ..........---.-.............(-(((....((((........(((((((((.((..((((((.....)))))).(.....)..........)))))))))))))))))))... ( -31.60) >DroEre_CAF1 18441 115 + 1 AAUAAC--AAGG-C-UGAAAAAAGGGAAC-CCCUAACGAGCGCUGGAAUUAUUUUUAGAGCAGUGCCGCAAAAAGUGGCAAAUAAUGCAAGUAUCUAAGCCUGCAAGUGGCGCGGGGAAA ......--....-.-.............(-(((......((.((((((....)))))).)).(((((((.....((.....))..((((.((......)).)))).)))))))))))... ( -31.20) >DroYak_CAF1 16371 120 + 1 AAUAAUACAAAGUGUACAAAAAAGCAAACCCCCCAACGAGCGCUGAAAUUAUUUUUAGAGCAGCGCCGCAAAAAGUGGCAAGUAAUGCAAAUAUCUAAGCCUGCAAGUGGCGCGGGGAAA ..............................((((.....((.((((((....)))))).)).(((((((.......(((...................))).....)))))))))))... ( -32.11) >DroAna_CAF1 40072 112 + 1 ---GGCACAG---A-GAAGGAAAAAAGAC-CCUUAACGAGCUCUAAAAUUAUUUUUAGAGCAGUGCCGCAAAAAGUGGCAAAUAAUGCAAAUAUCUAAGCAUGCAAGUGGCGCGAGGAAA ---(((((.(---.-.((((.........-))))..)..(((((((((....))))))))).))))).......(((.((....((((..........)))).....)).)))....... ( -33.10) >consensus AAUAACACAA___A_UAAAAAAAGGGAAC_CCCUAACGAGCGCUGAAAUCAUUUGUAGAGCAGUGCCGCAAAAAGUGGCAAGUAAUGCAAAUAUCUAAGCCUGCAAGUGGCGCGGGGAAA ..............................((((.....((((((...((.......)).))))(((((.....))))).......))..........(((.......)))..))))... (-21.88 = -21.47 + -0.41)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:28 2006