| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,959,345 – 1,959,447 |
| Length | 102 |
| Max. P | 0.681937 |

| Location | 1,959,345 – 1,959,447 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -23.73 |
| Consensus MFE | -17.26 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
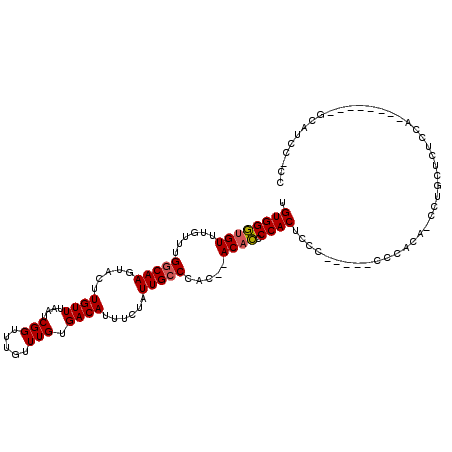
>2L_DroMel_CAF1 1959345 102 + 22407834 UGUGGGUGUUUGUUUGGCAAGUACUUGUUUAAUCGGUUUGUUUGUGACAUUUCUAUUGCCCAC--ACACCCCACUCCC-----CCCACAUCCUGCUCUGCA--------GCAUCC-CC .(((((.((.(((..(((((.....((((....(((.....))).))))......))))).))--).)))))))....-----........((((...)))--------).....-.. ( -23.40) >DroSec_CAF1 1847 102 + 1 UGUGGGUGUUUGUUUGGCAAGUACUUGUUUAAUCGGUUUGUUUGUGACAUUUCUAUUGCCCAC--ACACCCCACUCCC-----CCCACAUCCUACUCUCCA--------GCAUCC-CC .(((((.((.(((..(((((.....((((....(((.....))).))))......))))).))--).)))))))....-----..................--------......-.. ( -20.80) >DroSim_CAF1 5632 102 + 1 UGUGGGUGUUUGUUUGGCAAGUACUUGUUUAAUCGGUUUGUUUGUGACAUUUCUAUUGCCCAC--ACACCCCACUCCA-----CCCACAGCCUGCUCUCCA--------GCAUCA-CC ((((((((...(((..((((....))))..))).((..(((.((.(.((.......)).))).--)))..))....))-----))))))...((((....)--------)))...-.. ( -24.30) >DroEre_CAF1 11099 115 + 1 UGUGGGUGUUUGUUUGGCAAGUACUUGUUUAAUCGGUUUGUUUGUGACAUUUCUAUUGCCCAC--ACACCCCACAUCCCACAGCCCACAGCCAGCUUUCCGGGCUUACUGCAUCC-CC ((((((.((.(((..(((((.....((((....(((.....))).))))......))))).))--).))))))))......(((((..((....))....)))))..........-.. ( -29.00) >DroYak_CAF1 8905 85 + 1 UGUGGCUGUUUGUUUGGCAAGUACUUGUUUAAUCGGUUUGUUUGUGACAUUUCUAUUGCCCAC--ACAGCCCACAUCUCAC------------------------------AUCC-CC (((((((((.((...(((((.....((((....(((.....))).))))......))))))).--)))).)))))......------------------------------....-.. ( -20.40) >DroAna_CAF1 31495 101 + 1 UGUGGGCGUUUGUUUGCCAAGUACUUGUUUAAUCGGUUUGUUUGUGACAUUUCUAUUGCCCACCCACACCCCACUACA-----CCCCGA----ACGCCGGG--------GCAUCCCCC ....((((((((.....(((....))).......((..(((..(((.((.......))..)))..)))..))......-----...)))----)))))(((--------(...)))). ( -24.50) >consensus UGUGGGUGUUUGUUUGGCAAGUACUUGUUUAAUCGGUUUGUUUGUGACAUUUCUAUUGCCCAC__ACACCCCACUCCC_____CCCACA_CCUGCUCUCCA________GCAUCC_CC .((((((((......(((((.....((((....(((.....))).))))......))))).....)))).))))............................................ (-17.26 = -17.32 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:42:10 2006