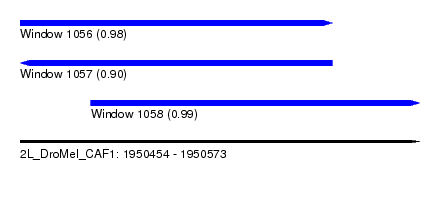
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,950,454 – 1,950,573 |
| Length | 119 |
| Max. P | 0.989084 |

| Location | 1,950,454 – 1,950,547 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 74.97 |
| Mean single sequence MFE | -26.41 |
| Consensus MFE | -18.05 |
| Energy contribution | -18.88 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1950454 93 + 22407834 AA--------GCUGUAACGCAAUGUUAAAAAC--ACCUUUUUCUAUAUCGAAACGUGUCCUGGAGGAUCUGUGUAGCG-UGUCUACACGGAACUCAGGGCUUUC--- ((--------((..((((.....))))...((--((...((((......)))).))))((((.((..(((((((((..-...))))))))).))))))))))..--- ( -24.30) >DroPse_CAF1 150907 101 + 1 AACAGAUGAUGCUCGUAGACAAUUCCGAAAAC-----AUUUCCUAUAUCAAAGCGCACUUCAGAGGAUCUGUGUAGCC-GAUCUACACAGAACUCAGUGCGUGGUGG ...((..((((.(((..........)))...)-----)))..))....((..(((((((...(((..(((((((((..-...))))))))).))))))))))..)). ( -30.10) >DroSec_CAF1 115121 93 + 1 AA--------GCUCUAACGGAAUAUUAAAAAC--ACCAUUUCCUAUAUCGAAACGAGACCUGGAGGAUCCGUGUAGCC-GAUCUACACGGAACUCAGGGCUUUC--- ((--------(((((...((((..........--.....))))...........(((.((....)).(((((((((..-...))))))))).))))))))))..--- ( -27.86) >DroSim_CAF1 99109 93 + 1 AA--------GCUCUAACGCAAAGUUAAAAAC--ACCAUUUCCUAUAUCGAAACGAGACCUGGAGGAUCCGUGUAGCC-GAUGUACACGGAACUCAGGGCUUUC--- ((--------(((((.................--....(((((....(((...))).....))))).((((((((...-....))))))))....)))))))..--- ( -23.10) >DroYak_CAF1 126400 96 + 1 AA--------GCUCUAACGCAAUGUUAAAAACACACCAUUUCCUAUAUCGAAACGAGUCCUGGAGGAUCCGUGUAGUUUGAUCUACACAGAACUCAGGGCUUUC--- ..--------((......))..................................((((((((.((..((.((((((......)))))).)).))))))))))..--- ( -23.00) >DroPer_CAF1 149546 101 + 1 AACAGAUGAUGCUCGUAGACAAUUCCGAAAAC-----AUUUCCUAUAUCAAAGCGCACUUCAGAGGAUCUGUGUAGCC-GAUCUACACAGAACUCAGUGCGUGGUGG ...((..((((.(((..........)))...)-----)))..))....((..(((((((...(((..(((((((((..-...))))))))).))))))))))..)). ( -30.10) >consensus AA________GCUCUAACGCAAUGUUAAAAAC__ACCAUUUCCUAUAUCGAAACGAGACCUGGAGGAUCCGUGUAGCC_GAUCUACACAGAACUCAGGGCUUUC___ ......................................................(((((((.(((..(((((((((......))))))))).))))))))))..... (-18.05 = -18.88 + 0.83)


| Location | 1,950,454 – 1,950,547 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 74.97 |
| Mean single sequence MFE | -27.87 |
| Consensus MFE | -17.05 |
| Energy contribution | -17.30 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.903438 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1950454 93 - 22407834 ---GAAAGCCCUGAGUUCCGUGUAGACA-CGCUACACAGAUCCUCCAGGACACGUUUCGAUAUAGAAAAAGGU--GUUUUUAACAUUGCGUUACAGC--------UU ---..((((((((((.((.((((((...-..)))))).))..)).))))((((.((((......))))...))--))...((((.....))))..))--------)) ( -22.60) >DroPse_CAF1 150907 101 - 1 CCACCACGCACUGAGUUCUGUGUAGAUC-GGCUACACAGAUCCUCUGAAGUGCGCUUUGAUAUAGGAAAU-----GUUUUCGGAAUUGUCUACGAGCAUCAUCUGUU ......(((((((((.(((((((((...-..)))))))))..)))...)))))).......(((((..((-----((((..(((....)))..))))))..))))). ( -31.10) >DroSec_CAF1 115121 93 - 1 ---GAAAGCCCUGAGUUCCGUGUAGAUC-GGCUACACGGAUCCUCCAGGUCUCGUUUCGAUAUAGGAAAUGGU--GUUUUUAAUAUUCCGUUAGAGC--------UU ---..((((((((((.(((((((((...-..)))))))))..)).))))..(((((((.......))))))).--...((((((.....))))))))--------)) ( -27.60) >DroSim_CAF1 99109 93 - 1 ---GAAAGCCCUGAGUUCCGUGUACAUC-GGCUACACGGAUCCUCCAGGUCUCGUUUCGAUAUAGGAAAUGGU--GUUUUUAACUUUGCGUUAGAGC--------UU ---..((((((((((.((((((((....-...))))))))..)).))))..(((((((.......))))))).--...((((((.....))))))))--------)) ( -26.00) >DroYak_CAF1 126400 96 - 1 ---GAAAGCCCUGAGUUCUGUGUAGAUCAAACUACACGGAUCCUCCAGGACUCGUUUCGAUAUAGGAAAUGGUGUGUUUUUAACAUUGCGUUAGAGC--------UU ---..((((((((((.(((((((((......)))))))))..)).))))(((((((((.......))))))).))...((((((.....))))))))--------)) ( -28.80) >DroPer_CAF1 149546 101 - 1 CCACCACGCACUGAGUUCUGUGUAGAUC-GGCUACACAGAUCCUCUGAAGUGCGCUUUGAUAUAGGAAAU-----GUUUUCGGAAUUGUCUACGAGCAUCAUCUGUU ......(((((((((.(((((((((...-..)))))))))..)))...)))))).......(((((..((-----((((..(((....)))..))))))..))))). ( -31.10) >consensus ___GAAAGCCCUGAGUUCCGUGUAGAUC_GGCUACACAGAUCCUCCAGGACUCGUUUCGAUAUAGGAAAUGGU__GUUUUUAACAUUGCGUUAGAGC________UU .......((((((((.(((((((((......)))))))))..))).)))...............((((((.....))))))..............)).......... (-17.05 = -17.30 + 0.25)

| Location | 1,950,475 – 1,950,573 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 78.18 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -20.19 |
| Energy contribution | -21.05 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.15 |
| SVM RNA-class probability | 0.989084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1950475 98 + 22407834 AAC--ACCUUUUUCUAUAUCGAAACGUGUCCUGGAGGAUCUGUGUAGCG-UGUCUACACGGAACUCAGGGCUUUC---GGUAU-UGGCGAC-----UAACUAGGUAAAUC ...--((((((.((.(((((((((...((((((.((..(((((((((..-...))))))))).))))))))))))---)))))-.)).)).-----.....))))..... ( -32.00) >DroPse_CAF1 150936 102 + 1 AAC-----AUUUCCUAUAUCAAAGCGCACUUCAGAGGAUCUGUGUAGCC-GAUCUACACAGAACUCAGUGCGUGGUGGGGUA--GGGAGACAUUCAGAACUAUCUAGAUC ...-----.((((((((..((..(((((((...(((..(((((((((..-...))))))))).))))))))))..))..)))--)))))....(((((....))).)).. ( -38.90) >DroSec_CAF1 115142 98 + 1 AAC--ACCAUUUCCUAUAUCGAAACGAGACCUGGAGGAUCCGUGUAGCC-GAUCUACACGGAACUCAGGGCUUUC---GGUAU-UGGCGAC-----UAACUAGUUAAAUC ...--.......((.(((((((((.(...((((.((..(((((((((..-...))))))))).)))))).)))))---)))))-.)).(((-----(....))))..... ( -32.70) >DroSim_CAF1 99130 98 + 1 AAC--ACCAUUUCCUAUAUCGAAACGAGACCUGGAGGAUCCGUGUAGCC-GAUGUACACGGAACUCAGGGCUUUC---GGUAU-UGGCGAC-----UAACUAGGUAAAUC ...--(((....((.(((((((((.(...((((.((..((((((((...-....)))))))).)))))).)))))---)))))-.)).(..-----...)..)))..... ( -29.30) >DroYak_CAF1 126421 102 + 1 AACACACCAUUUCCUAUAUCGAAACGAGUCCUGGAGGAUCCGUGUAGUUUGAUCUACACAGAACUCAGGGCUUUC---GCUAUUUGGCGAC-----UAACUAGGUAAAUC ........((((((((.........((((((((.((..((.((((((......)))))).)).))))))))))((---(((....))))).-----....)))).)))). ( -30.80) >DroPer_CAF1 149575 102 + 1 AAC-----AUUUCCUAUAUCAAAGCGCACUUCAGAGGAUCUGUGUAGCC-GAUCUACACAGAACUCAGUGCGUGGUGGGGUA--GGGAGACAUUCAGAACUAUCUAGAUC ...-----.((((((((..((..(((((((...(((..(((((((((..-...))))))))).))))))))))..))..)))--)))))....(((((....))).)).. ( -38.90) >consensus AAC__ACCAUUUCCUAUAUCGAAACGAGACCUGGAGGAUCCGUGUAGCC_GAUCUACACAGAACUCAGGGCUUUC___GGUAU_UGGCGAC_____UAACUAGGUAAAUC ............((..((((.....(((((((.(((..(((((((((......))))))))).)))))))))).....))))...))....................... (-20.19 = -21.05 + 0.86)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:51 2006