| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,944,085 – 1,944,202 |
| Length | 117 |
| Max. P | 0.875684 |

| Location | 1,944,085 – 1,944,202 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
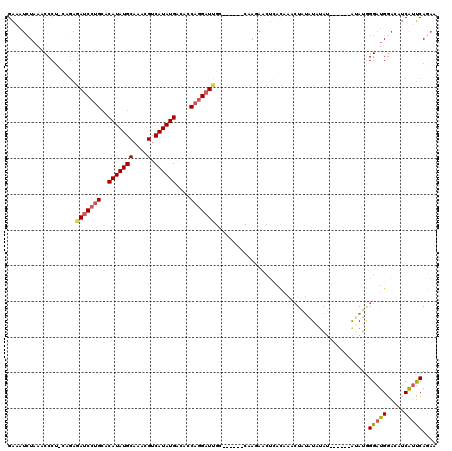
| Mean pairwise identity | 73.61 |
| Mean single sequence MFE | -25.15 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.25 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.79 |
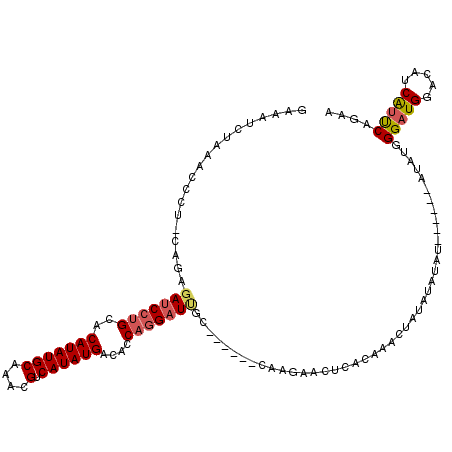
| Structure conservation index | 0.55 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875684 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1944085 117 + 22407834 GAAAUCUAAACCCU-CAGAGAUCCUGCACAUAUGCAAACGUCAUAUGACACCAGGAUAGAAUGACACCAGAACUCACAAACUUUAUAUAU--AUAUGUAUGGGAUGGACUUCAUUCAGAA ....(((.......-.....((((((..(((((((....).))))))....)))))).((((((..(((...((((.......(((((..--..))))))))).)))...))))))))). ( -23.51) >DroPse_CAF1 141484 107 + 1 GGAGCCUUAACCCUGGAGAGA-CCUGGCCAUAUGCAGCCGGCAUAUGGCUACUGGAUCGA------CACG------UAGCGUGUGUAUAUAUCUUAGCAUAGGAUGGGUAGCGGCCAACA .(.(((...((((......((-(((((((((((((.....)))))))))))..)).)).(------((((------...)))))......((((((...))))))))))...)))).... ( -35.10) >DroSec_CAF1 108780 107 + 1 GAAAUCUAAACCCU-CAGAGAUCCUGCACAUAUGCAAACGUCAUAUGACACCAGGAUUGC------CAAGAACUCACAAACUAUAUAUAU------AUAUGGGAUGCACUUCAUUCAGAA (((...........-....(((((((..(((((((....).))))))....)))))))..------...(((....((..((((((....------))))))..))...))).))).... ( -19.30) >DroSim_CAF1 92718 107 + 1 GAAAUCUAAACCCU-CAGAGAUCCUGCACAUAUGCAAACGUCAUAUGAUACCAGGAUUGC------CAAGAACUCACAAACUAUAUAUAU------AUAUGGGAUGGACUUCAUUCAGAA (((...........-....(((((((..(((((((....).))))))....)))))))..------...(((.((.((..((((((....------))))))..)))).))).))).... ( -22.90) >DroEre_CAF1 110949 107 + 1 GACAUCUAAUCCCU-CAAAGAUCCUGCACAUAUGCAAACGUCAUAUGGCAGCAGGAUUGC------CGAUAGCUUACUCACUAGAUUUAU------AGAUGGGAUGGGCAUCAUUCAGAA ....(((.......-....((((((((.(((((((....).))))))...))))))))..------.(((.((((((((((((......)------)).)))).))))))))....))). ( -31.20) >DroYak_CAF1 119360 104 + 1 GAAAUAUAAUCCCC-GAGAGAUC-UGCCCAUAUGCAAAAGUCAUAUGGCACCAGGAUUGC------CUACAACUCACUAACUAUAU--AU------AUAUUGGAUGGGCAUCAUUCAAAA ......((((((.(-....)...-((((.((((((....).)))))))))...)))))).------....................--..------...(((((((.....))))))).. ( -18.90) >consensus GAAAUCUAAACCCU_CAGAGAUCCUGCACAUAUGCAAACGUCAUAUGACACCAGGAUUGC______CAAGAACUCACAAACUAUAUAUAU______AUAUGGGAUGGACAUCAUUCAGAA ...................(((((((..(((((((....).))))))....)))))))...........................................(((((.....))))).... (-13.80 = -14.25 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:45 2006