| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,863,558 – 1,863,662 |
| Length | 104 |
| Max. P | 0.952317 |

| Location | 1,863,558 – 1,863,662 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.57 |
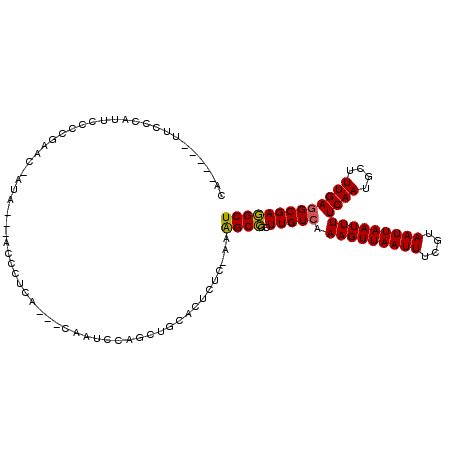
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -13.11 |
| Energy contribution | -12.87 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
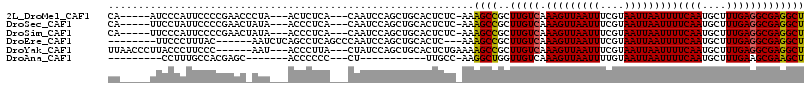
>2L_DroMel_CAF1 1863558 104 - 22407834 CA-----AUCCCAUUCCCCGAACCCUA---ACUCUCA---CAAUCCAGCUGCACUCUC-AAAGCCGCUUGUCAAAGUUAAUUUCGUAAUUAAUUUUCAAUGCUUUGAGGCGAGGCU ..-----....................---.......---......((((.(.(.(((-(((((...(((..((((((((((....))))))))))))).))))))))).).)))) ( -20.50) >DroSec_CAF1 31702 104 - 1 CA-----UUCCUAUUCCCCGAACUAUA---ACCCUCA---CAAUCCAGCUGCACUCUC-AAAGCCGCUUGUCAAAGUUAAUUUCGUAAUUAAUUUUCAAUGCUUUGAGGCGAGGCU ..-----....................---.......---......((((.(.(.(((-(((((...(((..((((((((((....))))))))))))).))))))))).).)))) ( -20.50) >DroSim_CAF1 32101 104 - 1 CA-----UUCCCAUUCCCCGAACUAUA---ACCCUCA---CAAUCCAGCUGCACUCUC-AAAGCCGCUUGUCAAAGUUAAUUUCGUAAUUAAUUUUCAAUGCUUUGAGGCGAGGCU ..-----....................---.......---......((((.(.(.(((-(((((...(((..((((((((((....))))))))))))).))))))))).).)))) ( -20.50) >DroEre_CAF1 31890 99 - 1 --------UUCCCUUUAC------AAUCUCAGCCUCAGCCCAAUCCAGCUGCACUC---AAAGCCGCUUGUCAAAGUUAAUUUCGUAAUUAAUUUUCAAUGCUUUGAGGCGAGGCU --------..........------......(((((((((........)))((.(((---(((((...(((..((((((((((....))))))))))))).)))))))))))))))) ( -30.80) >DroYak_CAF1 32638 104 - 1 UUAACCCUUACCCUUCCC------AAU---ACCCUUA---CUAUCCAGCUGCACUCUGAAAAGCCGCUUGUCAAAGUUAAUUUCGUAAUUAAUUUUCAAUGCUUUGAGGCGAGGCU ..................------...---.......---......((((((.(((...(((((...(((..((((((((((....))))))))))))).))))))))))..)))) ( -15.50) >DroAna_CAF1 34660 85 - 1 ---------CCUUUGCCACGAGC-------ACCCCCC---CU-----------UUGCC-AAGGCUGGUUGUCAAAGUUAAUUUUGUAAUUAAUUUUCAAUGCUUUGAAGCGAAGCU ---------....(((.....))-------)......---((-----------(((((-(((((..((((..((((((((((....))))))))))))))))))))..)))))).. ( -19.10) >consensus CA_____UUCCCAUUCCCCGAAC_AUA___ACCCUCA___CAAUCCAGCUGCACUCUC_AAAGCCGCUUGUCAAAGUUAAUUUCGUAAUUAAUUUUCAAUGCUUUGAGGCGAGGCU .............................................................((((..(((((.(((((((((....)))))))))((((....))))))))))))) (-13.11 = -12.87 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:41:25 2006