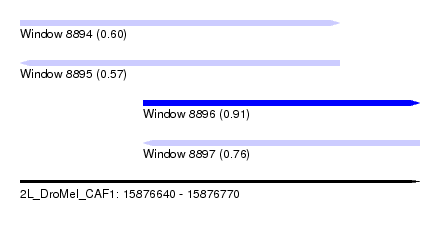
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 15,876,640 – 15,876,770 |
| Length | 130 |
| Max. P | 0.914768 |

| Location | 15,876,640 – 15,876,744 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -24.89 |
| Energy contribution | -25.50 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
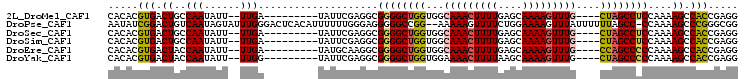
>2L_DroMel_CAF1 15876640 104 + 22407834 GAUGUGAGCGCUUAAAUUAUGUGCAAAAUGCUCGCAAAAGCCUCGGUGGCUUUUGGAGGCUAG----CAAACUUUUGCUCAAAAGUUUGCCACCAGCCCCGCCUCGAA ((.(((.((((.........)))).......((.((((((((.....))))))))))((((.(----(((((((((....))))))))))....)))).))).))... ( -36.10) >DroPse_CAF1 43124 105 + 1 GAUGUGUGCGCUUAAAUUAUGUGCAAAAUGCUCGCAAAAGCCGCCCGGGCUUUUGG-GGCUAAAAAAUAAACUUUUCCAGAAAACUUUUU--CCGGCCCCCUCCCAAA ......(((((.........)))))....((((.((((((((.....)))))))))-)))................((.((((....)))--).))............ ( -25.10) >DroSim_CAF1 50189 104 + 1 GAUGUGAGCGCUUAAAUUAUGUGCAAAAUGCUCGCAAAAGCCUCGGUGGCUUUUGGAGGCUAG----CAAACUUUUGCUCAAAAGUUUGCCACCAGCCCCGCCUCGAA ((.(((.((((.........)))).......((.((((((((.....))))))))))((((.(----(((((((((....))))))))))....)))).))).))... ( -36.10) >DroEre_CAF1 40204 104 + 1 GAUGUGAGCGCUUAAAUUAUGUGCAAAAUGCUCGCAAAAGCCUCGGUGGCUUUUGGGGGCUGG----CAAACUUUUGCUCAAAAGUUUGCCACCAGCCCCGCCUUGCA ..((..((.((.........(.((.....)).).((((((((.....))))))))((((((((----(((((((((....)))))))))))...))))))))))..)) ( -47.00) >DroYak_CAF1 26373 104 + 1 GAUGUGAGCGCUUAAAUUAUGUGCAAAAUGCUCGCAAAAGCCUCGGUGGCUUUUGGGGGCUAG----CAAACUUUUGCUUAAAAGUUUUCCACCAGCCCCGCCUCGAA ....((((.((.........(.((.....)).).((((((((.....))))))))((((((.(----.((((((((....))))))))....).)))))))))))).. ( -34.30) >DroPer_CAF1 69037 105 + 1 GAUGUGUGCGCUUAAAUUAUGUGCAAAAUGCUCGCAAAAGCCGCCCGGGCUUUUGG-GGCUAAAAAAUAAACUUUUCCAGAAAACUUUUU--CCGCCCCCCUCCCAAA ...((((((((.........)))))....((((.((((((((.....)))))))))-)))..............................--.)))............ ( -23.80) >consensus GAUGUGAGCGCUUAAAUUAUGUGCAAAAUGCUCGCAAAAGCCUCGGUGGCUUUUGG_GGCUAG____CAAACUUUUGCUCAAAAGUUUGCCACCAGCCCCGCCUCGAA ...((((((((...........)).....))))))(((((((.....)))))))((.((((.......((((((((....))))))))......))))))........ (-24.89 = -25.50 + 0.61)

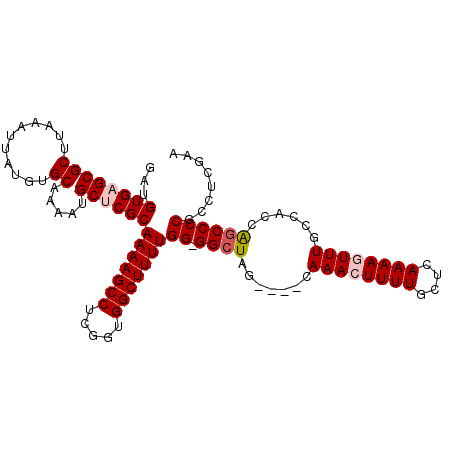
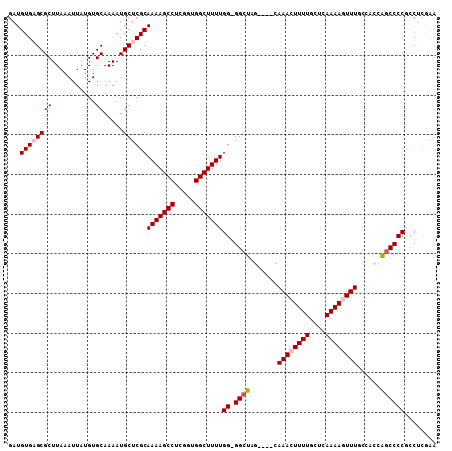
| Location | 15,876,640 – 15,876,744 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 85.69 |
| Mean single sequence MFE | -36.53 |
| Consensus MFE | -25.82 |
| Energy contribution | -26.85 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15876640 104 - 22407834 UUCGAGGCGGGGCUGGUGGCAAACUUUUGAGCAAAAGUUUG----CUAGCCUCCAAAAGCCACCGAGGCUUUUGCGAGCAUUUUGCACAUAAUUUAAGCGCUCACAUC ...(((((((((((...(((((((((((....)))))))))----))))))))((((((((.....))))))))...((.....))...........)).)))..... ( -39.30) >DroPse_CAF1 43124 105 - 1 UUUGGGAGGGGGCCGG--AAAAAGUUUUCUGGAAAAGUUUAUUUUUUAGCC-CCAAAAGCCCGGGCGGCUUUUGCGAGCAUUUUGCACAUAAUUUAAGCGCACACAUC .(((((..((((((((--(((....))))))((((((....)))))).)))-)).....)))))(((((....))..((.....))............)))....... ( -28.50) >DroSim_CAF1 50189 104 - 1 UUCGAGGCGGGGCUGGUGGCAAACUUUUGAGCAAAAGUUUG----CUAGCCUCCAAAAGCCACCGAGGCUUUUGCGAGCAUUUUGCACAUAAUUUAAGCGCUCACAUC ...(((((((((((...(((((((((((....)))))))))----))))))))((((((((.....))))))))...((.....))...........)).)))..... ( -39.30) >DroEre_CAF1 40204 104 - 1 UGCAAGGCGGGGCUGGUGGCAAACUUUUGAGCAAAAGUUUG----CCAGCCCCCAAAAGCCACCGAGGCUUUUGCGAGCAUUUUGCACAUAAUUUAAGCGCUCACAUC (((((.((((((((...(((((((((((....)))))))))----))))))))((((((((.....))))))))...))...)))))..................... ( -43.10) >DroYak_CAF1 26373 104 - 1 UUCGAGGCGGGGCUGGUGGAAAACUUUUAAGCAAAAGUUUG----CUAGCCCCCAAAAGCCACCGAGGCUUUUGCGAGCAUUUUGCACAUAAUUUAAGCGCUCACAUC ...((((((((((((((...((((((((....)))))))))----))))))))((((((((.....))))))))...((.....))...........)).)))..... ( -39.40) >DroPer_CAF1 69037 105 - 1 UUUGGGAGGGGGGCGG--AAAAAGUUUUCUGGAAAAGUUUAUUUUUUAGCC-CCAAAAGCCCGGGCGGCUUUUGCGAGCAUUUUGCACAUAAUUUAAGCGCACACAUC .(((((...(((((..--(((((((...((.....))...))))))).)))-)).....)))))(((((....))..((.....))............)))....... ( -29.60) >consensus UUCGAGGCGGGGCUGGUGGAAAACUUUUGAGCAAAAGUUUG____CUAGCC_CCAAAAGCCACCGAGGCUUUUGCGAGCAUUUUGCACAUAAUUUAAGCGCUCACAUC ........((((((((....((((((((....)))))))).....))))))))((((((((.....)))))))).((((.(((............))).))))..... (-25.82 = -26.85 + 1.03)
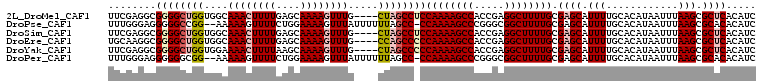

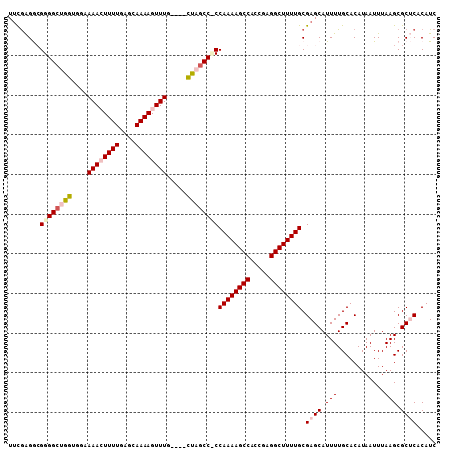
| Location | 15,876,680 – 15,876,770 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -29.33 |
| Consensus MFE | -22.94 |
| Energy contribution | -22.47 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914768 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15876680 90 + 22407834 CCUCGGUGGCUUUUGGAGGCUAG----CAAACUUUUGCUCAAAAGUUUGCCACCAGCCCCGCCUCGAAUA---------UCAA--AAUAUUGGCAGUCACGUGUG .....((((((...((.((((.(----(((((((((....))))))))))....))))))(((...((((---------(...--.))))))))))))))..... ( -30.60) >DroPse_CAF1 43164 102 + 1 CCGCCCGGGCUUUUGG-GGCUAAAAAAUAAACUUUUCCAGAAAACUUUUU--CCGGCCCCCUCCCAAAAAAUGUGAGUCCCAAAUACUAUUGACAGUCCGAUAUU .....((((((.((((-((((..((((...................))))--..))))))(((.((.....)).)))..............)).))))))..... ( -20.81) >DroSec_CAF1 50176 90 + 1 CCUCGGUGGCUUUUGGAGGCUAG----CAAACUUUUGCUCAAAAGUUUGCCACCAGCCCCGCCUCGAAUA---------UCAA--AAUAUUGGCAGUCACGUGUG .....((((((...((.((((.(----(((((((((....))))))))))....))))))(((...((((---------(...--.))))))))))))))..... ( -30.60) >DroSim_CAF1 50229 90 + 1 CCUCGGUGGCUUUUGGAGGCUAG----CAAACUUUUGCUCAAAAGUUUGCCACCAGCCCCGCCUCGAAUA---------UCAA--AAUAUUGGCAGUCACGUGUG .....((((((...((.((((.(----(((((((((....))))))))))....))))))(((...((((---------(...--.))))))))))))))..... ( -30.60) >DroEre_CAF1 40244 90 + 1 CCUCGGUGGCUUUUGGGGGCUGG----CAAACUUUUGCUCAAAAGUUUGCCACCAGCCCCGCCUUGCAUA---------UCAA--AAUAUUGGUAGUCACGUGUG .....(((((.....((((((((----(((((((((....)))))))))))...))))))........((---------((((--....)))))))))))..... ( -35.90) >DroYak_CAF1 26413 90 + 1 CCUCGGUGGCUUUUGGGGGCUAG----CAAACUUUUGCUUAAAAGUUUUCCACCAGCCCCGCCUCGAAUA---------CCAA--AAUAUUGGUAGUCACGUGUG ...(((.((((..(((..(((((----(((....)))))....)).)..)))..)))))))....((.((---------((((--....)))))).))....... ( -27.50) >consensus CCUCGGUGGCUUUUGGAGGCUAG____CAAACUUUUGCUCAAAAGUUUGCCACCAGCCCCGCCUCGAAUA_________UCAA__AAUAUUGGCAGUCACGUGUG .....((((((...((.((((......(((((((((....))))))))).....))))))(((............................)))))))))..... (-22.94 = -22.47 + -0.47)

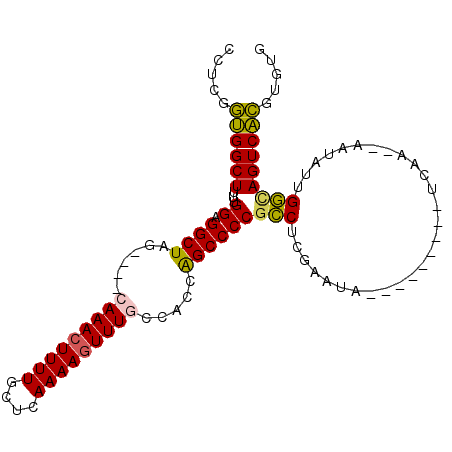
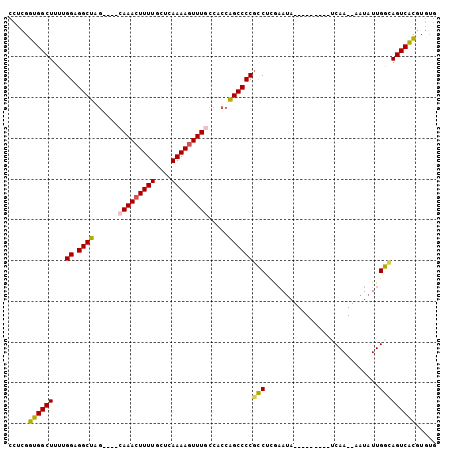
| Location | 15,876,680 – 15,876,770 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 81.82 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -21.15 |
| Energy contribution | -20.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 15876680 90 - 22407834 CACACGUGACUGCCAAUAUU--UUGA---------UAUUCGAGGCGGGGCUGGUGGCAAACUUUUGAGCAAAAGUUUG----CUAGCCUCCAAAAGCCACCGAGG .....(((.((((((((((.--...)---------))))...)))((((((...(((((((((((....)))))))))----)))))).))...)).)))..... ( -28.40) >DroPse_CAF1 43164 102 - 1 AAUAUCGGACUGUCAAUAGUAUUUGGGACUCACAUUUUUUGGGAGGGGGCCGG--AAAAAGUUUUCUGGAAAAGUUUAUUUUUUAGCC-CCAAAAGCCCGGGCGG .........(((((.........((((.(((.((.....)).)))((((((((--(((....))))))((((((....)))))).)))-)).....))))))))) ( -26.20) >DroSec_CAF1 50176 90 - 1 CACACGUGACUGCCAAUAUU--UUGA---------UAUUCGAGGCGGGGCUGGUGGCAAACUUUUGAGCAAAAGUUUG----CUAGCCUCCAAAAGCCACCGAGG .....(((.((((((((((.--...)---------))))...)))((((((...(((((((((((....)))))))))----)))))).))...)).)))..... ( -28.40) >DroSim_CAF1 50229 90 - 1 CACACGUGACUGCCAAUAUU--UUGA---------UAUUCGAGGCGGGGCUGGUGGCAAACUUUUGAGCAAAAGUUUG----CUAGCCUCCAAAAGCCACCGAGG .....(((.((((((((((.--...)---------))))...)))((((((...(((((((((((....)))))))))----)))))).))...)).)))..... ( -28.40) >DroEre_CAF1 40244 90 - 1 CACACGUGACUACCAAUAUU--UUGA---------UAUGCAAGGCGGGGCUGGUGGCAAACUUUUGAGCAAAAGUUUG----CCAGCCCCCAAAAGCCACCGAGG ....((.(.....(((....--))).---------.......(((((((((...(((((((((((....)))))))))----)))))))).....))).)))... ( -29.90) >DroYak_CAF1 26413 90 - 1 CACACGUGACUACCAAUAUU--UUGG---------UAUUCGAGGCGGGGCUGGUGGAAAACUUUUAAGCAAAAGUUUG----CUAGCCCCCAAAAGCCACCGAGG ....(((((.((((((....--))))---------)).))).((((((((((((...((((((((....)))))))))----)))))))).....)))..))... ( -33.10) >consensus CACACGUGACUGCCAAUAUU__UUGA_________UAUUCGAGGCGGGGCUGGUGGCAAACUUUUGAGCAAAAGUUUG____CUAGCCUCCAAAAGCCACCGAGG .....(((.((..(((......)))....................((((((((...(((((((((....)))))))))....))))))))....)).)))..... (-21.15 = -20.98 + -0.16)
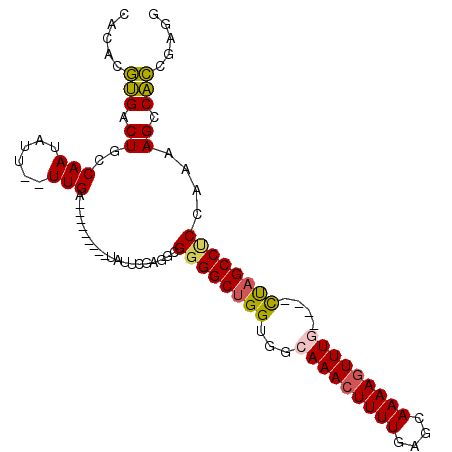
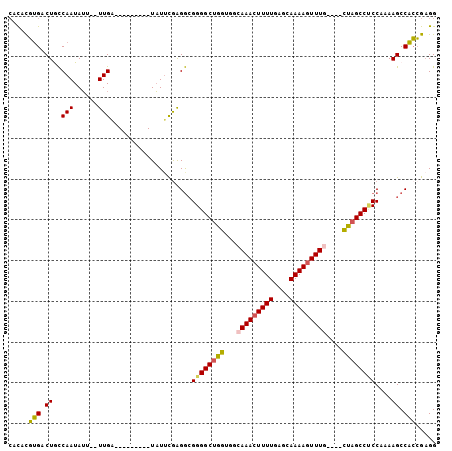
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:48:22 2006