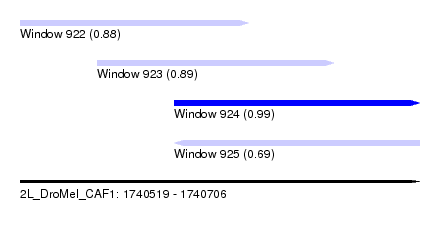
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,740,519 – 1,740,706 |
| Length | 187 |
| Max. P | 0.986284 |

| Location | 1,740,519 – 1,740,626 |
|---|---|
| Length | 107 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.48 |
| Mean single sequence MFE | -20.41 |
| Consensus MFE | -7.44 |
| Energy contribution | -8.62 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.36 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.876361 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1740519 107 + 22407834 AUUUUAAUAACUAUUUCCCAA----UAGUUACACACCUUUCCAUCAAGUGCAAACAACAUUCUAGGCGAUUCCUCC---------CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCC .......((((((((....))----))))))................((((((((((.....(((((.........---------..))))).))))))))))................. ( -18.70) >DroSim_CAF1 4119 107 + 1 AUUUCAAUAACUAUUUCCCAA----UAGUUACACACCUUUCCAUCAAGUGCAAACAACAUUCUAGGCGAUUCCUCC---------CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCC .......((((((((....))----))))))................((((((((((.....(((((.........---------..))))).))))))))))................. ( -18.70) >DroWil_CAF1 13312 112 + 1 A-UUUGAUGAUUUUAUGGCUAUAUUUAGUUACCUACUGU-----UAG--GUAAACAAAAUUGUUUAGCAUUCCAACACUAUUACUUAAAAUCUUAGUUUGCUUAAAUUGUUAGACCGUCC .-(((((..((((...(((((.((((..(((((((....-----)))--))))...))))....))))....(((.((((.............))))))))..))))..)))))...... ( -17.12) >DroYak_CAF1 4136 107 + 1 UGUUAAUAAACUAUUUCCCAA----UAGUUACACACCUGUCCAUCAAGUGCAAACAACAUUCUAGGCGAUUCCGCC---------CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCC (((((((.(((((((....))----))))).................((((((((((....((.((((....))))---------.)).....)))))))))).....)))))))..... ( -27.10) >consensus AUUUUAAUAACUAUUUCCCAA____UAGUUACACACCUUUCCAUCAAGUGCAAACAACAUUCUAGGCGAUUCCUCC_________CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCC ...............................................((((((((((.....(((((....................))))).))))))))))................. ( -7.44 = -8.62 + 1.19)


| Location | 1,740,555 – 1,740,666 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.74 |
| Mean single sequence MFE | -24.72 |
| Consensus MFE | -12.79 |
| Energy contribution | -15.10 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1740555 111 + 22407834 CCAUCAAGUGCAAACAACAUUCUAGGCGAUUCCUCC---------CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCCUAAAUCGGGCAGAAAAUUACUGACCUAAGUCAAGGGUACA ((.....((((((((((.....(((((.........---------..))))).)))))))))).(((((((..(...(((......)))..)..)))))))(((....)))..))..... ( -25.90) >DroSim_CAF1 4155 111 + 1 CCAUCAAGUGCAAACAACAUUCUAGGCGAUUCCUCC---------CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCCUAAAUCGGGCAGAAAAUUACUGACCUAAGUCAAGGGUACA ((.....((((((((((.....(((((.........---------..))))).)))))))))).(((((((..(...(((......)))..)..)))))))(((....)))..))..... ( -25.90) >DroWil_CAF1 13350 106 + 1 ----UAG--GUAAACAAAAUUGUUUAGCAUUCCAACACUAUUACUUAAAAUCUUAGUUUGCUUAAAUUGUUAGACCGUCCAAAAACAGGA--------ACUGACCUAACCCAAUGGGCAA ----..(--.((((((....)))))).).............................(((((((....(((((....(((.......)))--------......)))))....))))))) ( -16.40) >DroYak_CAF1 4172 111 + 1 CCAUCAAGUGCAAACAACAUUCUAGGCGAUUCCGCC---------CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCCUAAAUCGGGCAGAAAAUUACUGACCUAAGUCAAGGGUACA ((.....((((((((((....((.((((....))))---------.)).....)))))))))).(((((((..(...(((......)))..)..)))))))(((....)))..))..... ( -30.70) >consensus CCAUCAAGUGCAAACAACAUUCUAGGCGAUUCCUCC_________CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCCUAAAUCGGGCAGAAAAUUACUGACCUAAGUCAAGGGUACA .......((((((((((.....(((((....................))))).)))))))))).............(((((.....((((((.......))).)))......)))))... (-12.79 = -15.10 + 2.31)

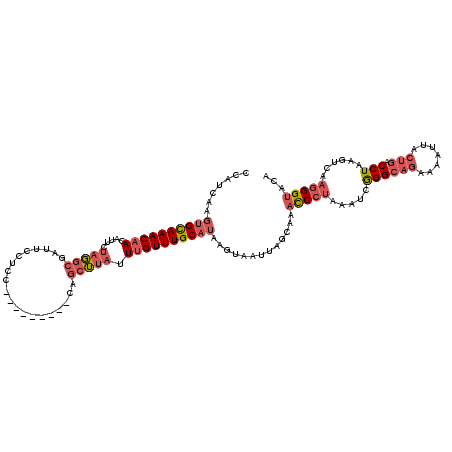
| Location | 1,740,591 – 1,740,706 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -22.58 |
| Consensus MFE | -14.96 |
| Energy contribution | -15.65 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986284 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1740591 115 + 22407834 -----CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCCUAAAUCGGGCAGAAAAUUACUGACCUAAGUCAAGGGUACAUAAAAUUAGCAAACAUUAGCUAACUGCGGAAUAAUCAACU -----.((((....(((((((.(((....)))....(((((...(..(((((.......))).))..)....)))))...........)))))))..))))................... ( -25.60) >DroSim_CAF1 4191 115 + 1 -----CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCCUAAAUCGGGCAGAAAAUUACUGACCUAAGUCAAGGGUACAUAAAAUUAGCAAACAUUAGCGAACUGCGGAAUAAUCAACU -----..(((....(((((((.(((....)))....(((((...(..(((((.......))).))..)....)))))...........)))))))..))).................... ( -24.90) >DroWil_CAF1 13384 98 + 1 UUACUUAAAAUCUUAGUUUGCUUAAAUUGUUAGACCGUCCAAAAACAGGA--------ACUGACCUAACCCAAUGGGCAAUAAAAUUAGCAAAUUCUAGUCAAUUC-------------- ...........((.((((((((......(((((....(((.......)))--------.)))))....(((...)))..........))))))))..)).......-------------- ( -14.20) >DroYak_CAF1 4208 115 + 1 -----CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCCUAAAUCGGGCAGAAAAUUACUGACCUAAGUCAAGGGUACAUAAAAUUAGCAAACAUUAGCUAACUGGAGAAUAAUCAACU -----.((((....(((((((.(((....)))....(((((...(..(((((.......))).))..)....)))))...........)))))))..))))................... ( -25.60) >consensus _____CAGCUUAUUUGUUUGCAUAAGUAAUUAGCAAACCCUAAAUCGGGCAGAAAAUUACUGACCUAAGUCAAGGGUACAUAAAAUUAGCAAACAUUAGCUAACUGCGGAAUAAUCAACU .......(((....(((((((.(((....(((....(((((.....((((((.......))).)))......)))))...)))..))))))))))..))).................... (-14.96 = -15.65 + 0.69)

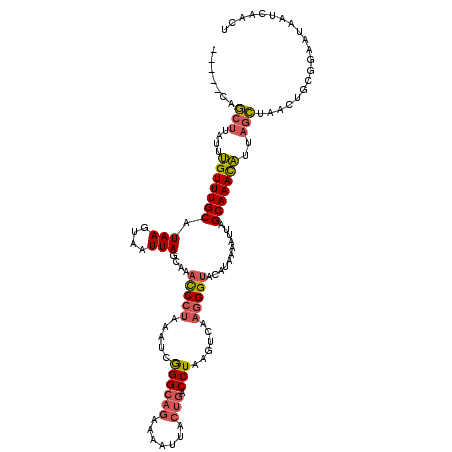
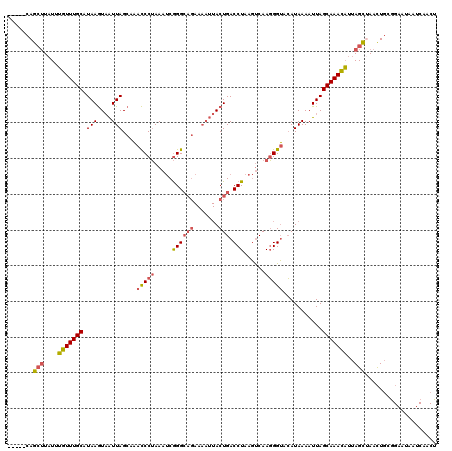
| Location | 1,740,591 – 1,740,706 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.04 |
| Mean single sequence MFE | -24.79 |
| Consensus MFE | -16.80 |
| Energy contribution | -17.93 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687159 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1740591 115 - 22407834 AGUUGAUUAUUCCGCAGUUAGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUUCUGCCCGAUUUAGGGUUUGCUAAUUACUUAUGCAAACAAAUAAGCUG----- .((.((....)).))...(((((..(((((((................((((....))))((((((....((((......)))).....))))))....)))))))....)))))----- ( -27.50) >DroSim_CAF1 4191 115 - 1 AGUUGAUUAUUCCGCAGUUCGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUUCUGCCCGAUUUAGGGUUUGCUAAUUACUUAUGCAAACAAAUAAGCUG----- .((.((....)).)).....(((..(((((((................((((....))))((((((....((((......)))).....))))))....)))))))....)))..----- ( -26.20) >DroWil_CAF1 13384 98 - 1 --------------GAAUUGACUAGAAUUUGCUAAUUUUAUUGCCCAUUGGGUUAGGUCAGU--------UCCUGUUUUUGGACGGUCUAACAAUUUAAGCAAACUAAGAUUUUAAGUAA --------------..............((((((((((((((((.......(((((..(.((--------((........)))))..))))).......))))..)))))))...))))) ( -18.34) >DroYak_CAF1 4208 115 - 1 AGUUGAUUAUUCUCCAGUUAGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUUCUGCCCGAUUUAGGGUUUGCUAAUUACUUAUGCAAACAAAUAAGCUG----- ((((((((.......))))))))..(((((((................((((....))))((((((....((((......)))).....))))))....))))))).........----- ( -27.10) >consensus AGUUGAUUAUUCCGCAGUUAGCUAAUGUUUGCUAAUUUUAUGUACCCUUGACUUAGGUCAGUAAUUUUCUGCCCGAUUUAGGGUUUGCUAAUUACUUAUGCAAACAAAUAAGCUG_____ ..................(((((..(((((((................((((....))))((((((....((((......)))).....))))))....)))))))....)))))..... (-16.80 = -17.93 + 1.13)

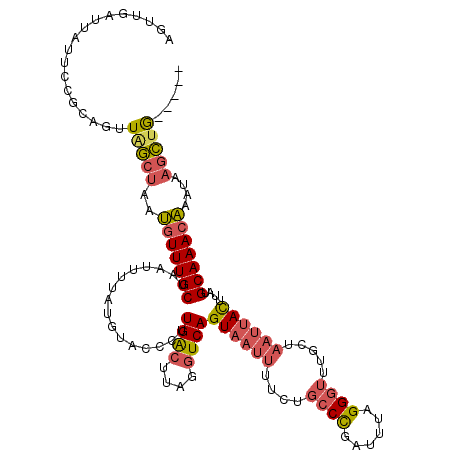
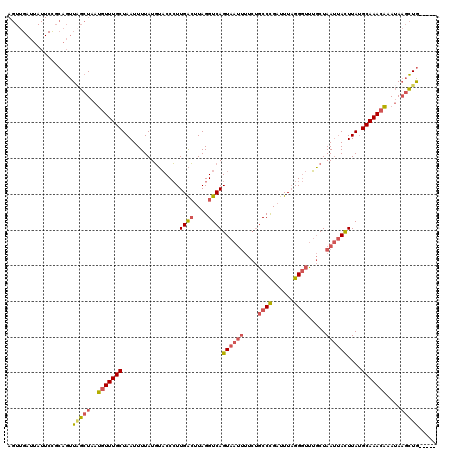
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:39:41 2006