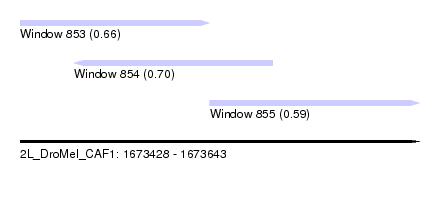
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,673,428 – 1,673,643 |
| Length | 215 |
| Max. P | 0.696673 |

| Location | 1,673,428 – 1,673,530 |
|---|---|
| Length | 102 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.01 |
| Mean single sequence MFE | -19.80 |
| Consensus MFE | -12.56 |
| Energy contribution | -13.12 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1673428 102 + 22407834 -----------ACACAUCAUUGUGGGCUGCAGCAGCAGCAGCAGCAACCCCGAACGAGAUUCAUUAAAAA-------AAUGUUGGGAAUAUAAAAAAUUGUUUUCCAUACGCAUUAAUGC -----------.((((....)))).(((((.((....)).))))).........................-------(((((..((((.((((....)))).))))....)))))..... ( -22.20) >DroSim_CAF1 799 95 + 1 ------AACUCACACA---UCGUGGGCUGCAG---------CAGCAACCCCGAACGAGAUUCAUUAAAAA-------AAUGUUGGGAAUAUAAAAAAUUGUUUUCCAUACGCAUUAAUGC ------..(((.....---(((.(((.(((..---------..))).))))))..)))............-------(((((..((((.((((....)))).))))....)))))..... ( -20.50) >DroYak_CAF1 819 108 + 1 ACACACAACACACACA---UCCUUUGCUGCAG---------CAGCAACCCCGAACGAGAUUCAUUAACAAAAAAUGAAAUGCUCGGAACAUAAAAAAUUGUUUUCCAUACGCAUUAAUGC ................---....((((((...---------))))))........(((.((((((.......))))))...)))((((.((((....)))).))))....(((....))) ( -16.70) >consensus ______AAC_CACACA___UCGUGGGCUGCAG_________CAGCAACCCCGAACGAGAUUCAUUAAAAA_______AAUGUUGGGAAUAUAAAAAAUUGUUUUCCAUACGCAUUAAUGC ...................(((.(((.(((.............))).))))))........................(((((..((((.((((....)))).))))....)))))..... (-12.56 = -13.12 + 0.56)

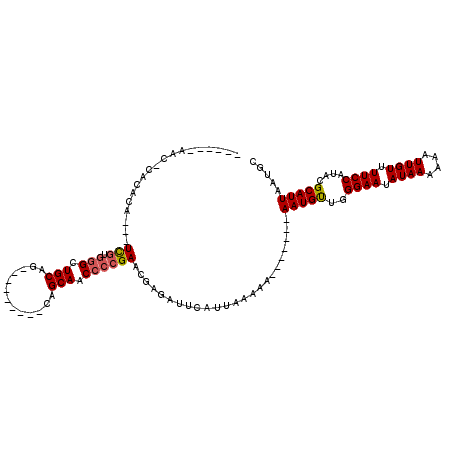

| Location | 1,673,457 – 1,673,564 |
|---|---|
| Length | 107 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.26 |
| Mean single sequence MFE | -25.25 |
| Consensus MFE | -19.37 |
| Energy contribution | -19.04 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696673 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1673457 107 - 22407834 UCCGUAACCACGUU------UACUUUACUUACGCAUUGCUGCAUUAAUGCGUAUGGAAAACAAUUUUUUAUAUUCCCAACAUU-------UUUUUAAUGAAUCUCGUUCGGGGUUGCUGC ...((((((..(((------(.((.....(((((((((......))))))))).)).)))).............((.(((...-------.(((....)))....))).))))))))... ( -22.50) >DroSim_CAF1 822 104 - 1 UCCUUAACCGCGUU--------CUUUACUUACGCAUUGCUGCAUUAAUGCGUAUGGAAAACAAUUUUUUAUAUUCCCAACAUU-------UUUUUAAUGAAUCUCGUUCGGGGUUGCUG- ....(((((.((..--------(......(((((((((......))))))))).((((..............))))...((((-------.....))))......)..)).)))))...- ( -19.94) >DroYak_CAF1 848 119 - 1 UCCGCGAUCACGUUGUUACGUUCUUUACUUACGCAUUGCUGCAUUAAUGCGUAUGGAAAACAAUUUUUUAUGUUCCGAGCAUUUCAUUUUUUGUUAAUGAAUCUCGUUCGGGGUUGCUG- ...((((((..((((((...((((.....(((((((((......))))))))).))))))))))..........((((((..((((((.......))))))....))))))))))))..- ( -33.30) >consensus UCCGUAACCACGUU______U_CUUUACUUACGCAUUGCUGCAUUAAUGCGUAUGGAAAACAAUUUUUUAUAUUCCCAACAUU_______UUUUUAAUGAAUCUCGUUCGGGGUUGCUG_ ...((((((.((.................(((((((((......))))))))).((((..............))))................................)).))))))... (-19.37 = -19.04 + -0.33)

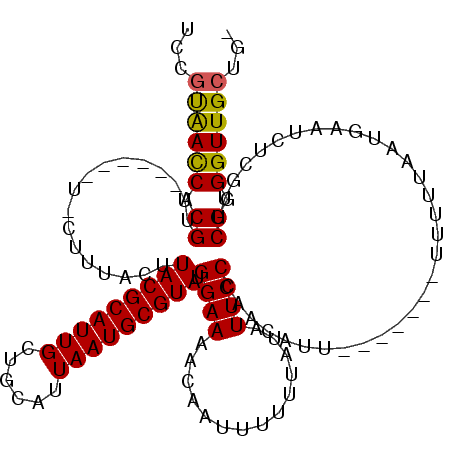
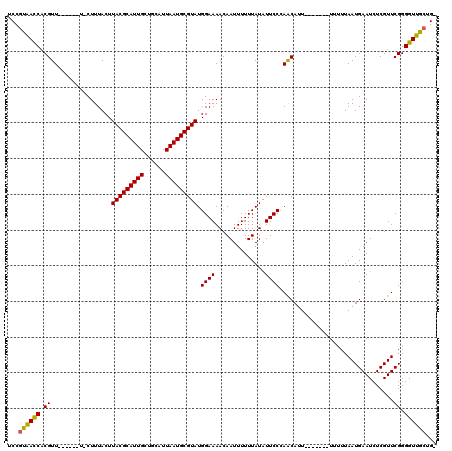
| Location | 1,673,530 – 1,673,643 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -32.05 |
| Consensus MFE | -26.16 |
| Energy contribution | -25.41 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.589809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1673530 113 + 22407834 AGCAAUGCGUAAGUAAAGUA------AACGUGGUUACGGAAUGGGAAAGUGUGGUGGCCAGACUUGGUUAUG-ACUAUAAAGCCAUGUGGCAUAACAUGGGGUAGUCCUCCUUGGGAUUU .....(((....)))..(((------(......))))(((((((..((((.(((...))).))))..))))(-(((((....((((((......)))))).)))))).)))......... ( -28.40) >DroSec_CAF1 5 111 + 1 AGCAAUGCGUAAGUAAAG--------AACGCGGUUAAGGAAUGGGAAGGUGUGGUGGCCAGACUUGGUUAUG-ACUAUAAAGCCAUGUGGCAUAACAUGGGGUAGUCCUCCUUGGGAUUU .....(((((........--------.))))).(((((((((((..((((.(((...))).))))..))))(-(((((....((((((......)))))).)))))).)))))))..... ( -31.80) >DroSim_CAF1 894 111 + 1 AGCAAUGCGUAAGUAAAG--------AACGCGGUUAAGGAAUGGGGAGGUGUGGUGGCCAGACUUGGUUAUG-ACUAUAAAGCCAUGUGGCAUAACAUGGGGUAGUCCUCCUUGGGAUUU .....(((((........--------.)))))..........((((((.....((((((......))))))(-(((((....((((((......)))))).))))))))))))....... ( -31.70) >DroYak_CAF1 927 120 + 1 AGCAAUGCGUAAGUAAAGAACGUAACAACGUGAUCGCGGAAUGGGAAAGUGUGGUGGCCAGACUUGGCUAUGGACUAUAAAGCCAUGUGGAAUAACAUGGGGUAGUCCUUCUUGGGAUUU .....(((....)))....((((....))))((((.((((((((..((((.(((...))).))))..))))(((((((....((((((......)))))).))))))).)).)).)))). ( -36.30) >consensus AGCAAUGCGUAAGUAAAG________AACGCGGUUAAGGAAUGGGAAAGUGUGGUGGCCAGACUUGGUUAUG_ACUAUAAAGCCAUGUGGCAUAACAUGGGGUAGUCCUCCUUGGGAUUU .....(((((.................))))).(((((((((((..((((.(((...))).))))..))))(.(((((....((((((......)))))).))))).))))))))..... (-26.16 = -25.41 + -0.75)

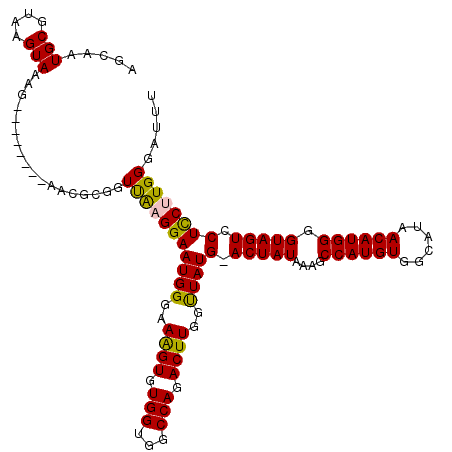
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:34 2006