
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,615,890 – 1,616,016 |
| Length | 126 |
| Max. P | 0.980415 |

| Location | 1,615,890 – 1,615,981 |
|---|---|
| Length | 91 |
| Sequences | 3 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 72.58 |
| Mean single sequence MFE | -15.77 |
| Consensus MFE | -11.39 |
| Energy contribution | -12.51 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.806381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
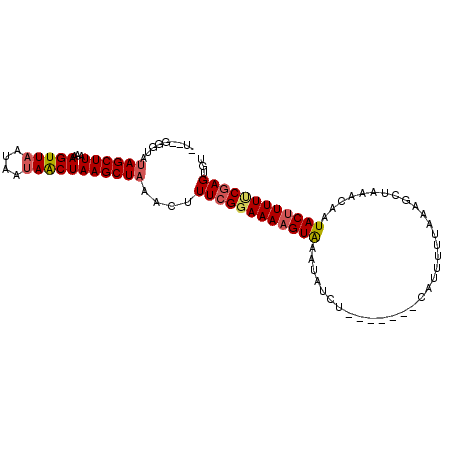
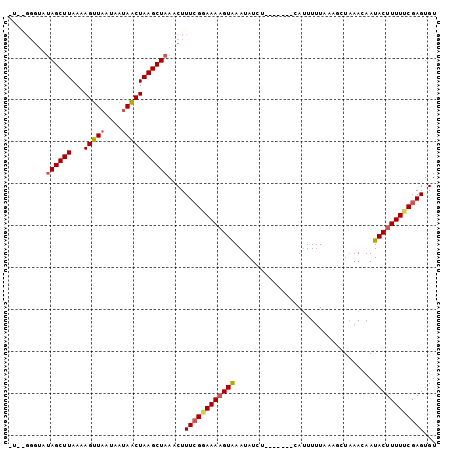
>2L_DroMel_CAF1 1615890 91 - 22407834 ----GGGUAUAGCUUAAAAGUUAAUAAUAACUAAGCUAAACUUUAGGAAAUGUAAAUAUCU-------CAAUUAUAAAGCUAAACAAUACUUUUUCGAGUGU ----((((.((((((...(((((....))))))))))).))))..................-------..................((((((....)))))) ( -11.80) >DroSec_CAF1 94347 95 - 1 AUGGGGGUGCAGCUUAAAAGUUAAUAAUAACUAAGCUAAUCUUUCGGAAAAGUGAAUAUCU-------UAUUUUUCAAGCUAAAGAAUACUUUUCCGAGUGU ...(((((..(((((...(((((....)))))))))).)))))(((((((((((....(((-------(.............)))).))))))))))).... ( -22.32) >DroYak_CAF1 95029 93 - 1 CUUA----UUAGCUUAAAAGUUAG---AAGCUAAGCUAACCUUUCGCAAAAGUAAAUAUCCCUAAAUACAUUUUUUA--CUUAACAAUACUUUUUCGAGUGU ....----(((((((...(((...---..))))))))))...((((.(((((((........((((.......))))--........))))))).))))... ( -13.19) >consensus _U__GGGUAUAGCUUAAAAGUUAAUAAUAACUAAGCUAAACUUUCGGAAAAGUAAAUAUCU_______CAUUUUUAAAGCUAAACAAUACUUUUUCGAGUGU .........((((((...(((((....)))))))))))....((((((((((((.................................))))))))))))... (-11.39 = -12.51 + 1.12)

| Location | 1,615,920 – 1,616,016 |
|---|---|
| Length | 96 |
| Sequences | 3 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 69.21 |
| Mean single sequence MFE | -21.67 |
| Consensus MFE | -12.96 |
| Energy contribution | -12.52 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
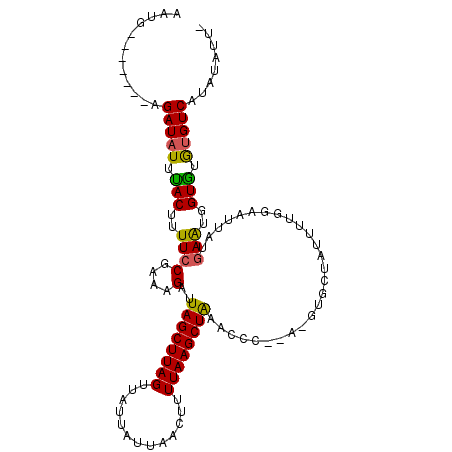
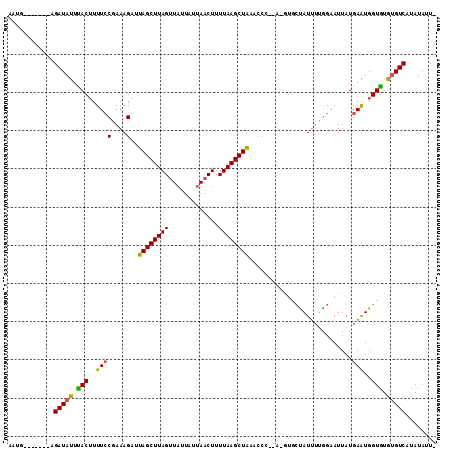
>2L_DroMel_CAF1 1615920 96 + 22407834 AUUG-------AGAUAUUUACAUUUCCUAAAGUUUAGCUUAGUUAUUAUUAACUUUUAAGCUAUACCC--------UAUUUGGGAAUUAUGAAUGGUGUGUGUCAUAUAUU- ..((-------(.((((...(((((((((((((.(((((((((((....)))))...)))))).))..--------..)))))))......))))..)))).)))......- ( -21.30) >DroSec_CAF1 94377 104 + 1 AAUA-------AGAUAUUCACUUUUCCGAAAGAUUAGCUUAGUUAUUAUUAACUUUUAAGCUGCACCCCCAUGUGCUAUGCUAGAAUUAUGAAUGGUGUGUGUCAUAUAUU- ....-------.(((((.((((.((((....).(((((..(((((....)))))........((((......))))...)))))......))).)))).))))).......- ( -22.90) >DroYak_CAF1 95057 105 + 1 AAUGUAUUUAGGGAUAUUUACUUUUGCGAAAGGUUAGCUUAGCUU---CUAACUUUUAAGCUAA----UAAGGUGCCAUUUUGGGAUUAUGAGUGGUAUAAGUCUUAUUUUC ..........(((((.....(((...(....)((((((((((...---.......)))))))))----))))(((((((((.........)))))))))..)))))...... ( -20.80) >consensus AAUG_______AGAUAUUUACUUUUCCGAAAGAUUAGCUUAGUUAUUAUUAACUUUUAAGCUAAACCC__A_GUGCUAUUUUGGAAUUAUGAAUGGUGUGUGUCAUAUAUU_ ............(((((.(((..((((....)..((((((((.............))))))))...........................)))..))).)))))........ (-12.96 = -12.52 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:25 2006