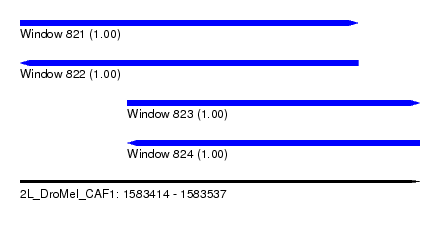
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,583,414 – 1,583,537 |
| Length | 123 |
| Max. P | 0.999957 |

| Location | 1,583,414 – 1,583,518 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 73.02 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -16.79 |
| Energy contribution | -20.96 |
| Covariance contribution | 4.17 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.18 |
| SVM RNA-class probability | 0.998678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1583414 104 + 22407834 UGACGUUUUUGGCACUCUGCUUAAAACUCGCUUUAGAAGC--------ACAGAGUGACCAUGCACCGCAACGGCAUUUCCGUCACUGUGCAAGGUAACACUGUCUUC-ACAAA .((.(((((.(((.....))).)))))))......((((.--------((((.((.(((.(((((.(..((((.....))))..).))))).))).)).))))))))-..... ( -34.90) >DroSec_CAF1 61912 104 + 1 UGACGUUUUCGGCACUCUGCUUAAAACUUGCCUUAGAAGC--------ACAGAGUGACCAUGCACCGGAACGGUAUUUCCGUCACUGUGCAAGGUCAUACUGUAUUC-ACAAU ....(((((.(((.....))).)))))........(((..--------((((.((((((.(((((.(..((((.....))))..).))))).)))))).)))).)))-..... ( -38.60) >DroSim_CAF1 64204 104 + 1 UGACGUUUUUGGAACUCUGCUUAAAACUUGGCUUAGAAGC--------ACAGAGUGAGCAUGCACCGCAACGGAAUUUCCGUCACUGUGCAAGGUCAUACUGUUUUC-ACAAU ........(((.......(((........)))...((((.--------((((.((((.(.(((((.(..((((.....))))..).))))).).)))).))))))))-.))). ( -29.20) >DroEre_CAF1 63082 101 + 1 UCACGUUUUUGGCACUUUGCCUAAAACUUGCUUCUAAAGCACGAGGGCACAGAGUGACCAUGC-----------AAUUCCGCCACUGUGCAAGGUCACACUAUCUUGCAUAA- ....((...(((((((((((((....(.((((.....)))).).)))))..))))).))).))-----------...........((((((((((......)))))))))).- ( -29.70) >DroYak_CAF1 62046 85 + 1 -------------GCUCUGCCUAA-AUUUGCUUUCAAAGC--------ACAGAGUUACCAUGCUUCCCUACGGUAUUUCAGCCACUGUGCAAGGUCACACUGUCUUG------ -------------.....((((..-.((((....))))((--------(((((((......))).......(((......))).)))))).))))............------ ( -16.70) >consensus UGACGUUUUUGGCACUCUGCUUAAAACUUGCUUUAGAAGC________ACAGAGUGACCAUGCACCGCAACGGUAUUUCCGUCACUGUGCAAGGUCACACUGUCUUC_ACAA_ ....(((((.(((.....))).))))).....................((((.((((((.(((((.(..((((.....))))..).))))).)))))).)))).......... (-16.79 = -20.96 + 4.17)

| Location | 1,583,414 – 1,583,518 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 73.02 |
| Mean single sequence MFE | -31.93 |
| Consensus MFE | -20.68 |
| Energy contribution | -24.24 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1583414 104 - 22407834 UUUGU-GAAGACAGUGUUACCUUGCACAGUGACGGAAAUGCCGUUGCGGUGCAUGGUCACUCUGU--------GCUUCUAAAGCGAGUUUUAAGCAGAGUGCCAAAAACGUCA ((((.-((((((((.((.(((.(((((.(..((((.....))))..).))))).))).)).))))--------.))))))))..(((((((..((.....))..))))).)). ( -39.20) >DroSec_CAF1 61912 104 - 1 AUUGU-GAAUACAGUAUGACCUUGCACAGUGACGGAAAUACCGUUCCGGUGCAUGGUCACUCUGU--------GCUUCUAAGGCAAGUUUUAAGCAGAGUGCCGAAAACGUCA .....-((((((((..(((((.(((((.(..((((.....))))..).))))).)))))..))))--------).)))...(((..(((((..((.....))..)))))))). ( -37.90) >DroSim_CAF1 64204 104 - 1 AUUGU-GAAAACAGUAUGACCUUGCACAGUGACGGAAAUUCCGUUGCGGUGCAUGCUCACUCUGU--------GCUUCUAAGCCAAGUUUUAAGCAGAGUUCCAAAAACGUCA (((((-....))))).((((..(((((.(..((((.....))))..).))))).....(((((((--------(((....)))..........))))))).........)))) ( -32.90) >DroEre_CAF1 63082 101 - 1 -UUAUGCAAGAUAGUGUGACCUUGCACAGUGGCGGAAUU-----------GCAUGGUCACUCUGUGCCCUCGUGCUUUAGAAGCAAGUUUUAGGCAAAGUGCCAAAAACGUGA -...(((...((((.((((((.((((..((......)))-----------))).)))))).))))((......)).......))).(((((.(((.....))).))))).... ( -29.70) >DroYak_CAF1 62046 85 - 1 ------CAAGACAGUGUGACCUUGCACAGUGGCUGAAAUACCGUAGGGAAGCAUGGUAACUCUGU--------GCUUUGAAAGCAAAU-UUAGGCAGAGC------------- ------....((((.((.(((.(((.(((...))).....((....))..))).))).)).))))--------((((((.........-.....))))))------------- ( -19.94) >consensus _UUGU_GAAGACAGUGUGACCUUGCACAGUGACGGAAAUACCGUUGCGGUGCAUGGUCACUCUGU________GCUUCUAAAGCAAGUUUUAAGCAGAGUGCCAAAAACGUCA ..........((((.((((((.(((((.(((((((.....))))))).))))).)))))).))))........((((..((......))..)))).................. (-20.68 = -24.24 + 3.56)

| Location | 1,583,447 – 1,583,537 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 71.01 |
| Mean single sequence MFE | -27.61 |
| Consensus MFE | -22.21 |
| Energy contribution | -23.90 |
| Covariance contribution | 1.69 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.80 |
| SVM decision value | 4.46 |
| SVM RNA-class probability | 0.999903 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1583447 90 + 22407834 UAGAAGCACAGAGUGACCAUGCACCGCAACGGCAUUUCCGUCACUGUGCAAGGUAACACUGUCUUCACAAAUAA-CUCG-------------------AUUUUUCACUUU ..((((.((((.((.(((.(((((.(..((((.....))))..).))))).))).)).))))))))........-....-------------------............ ( -27.60) >DroSec_CAF1 61945 110 + 1 UAGAAGCACAGAGUGACCAUGCACCGGAACGGUAUUUCCGUCACUGUGCAAGGUCAUACUGUAUUCACAAUCAAAAUCGAUGUCGCGUGCUUAUAAGUAUUUUUCACUAU ..(((((((((.((((((.(((((.(..((((.....))))..).))))).)))))).)))).......(((......)))...))(((((....)))))..)))..... ( -35.10) >DroSim_CAF1 64237 110 + 1 UAGAAGCACAGAGUGAGCAUGCACCGCAACGGAAUUUCCGUCACUGUGCAAGGUCAUACUGUUUUCACAAUCAAAAUCGAUGUCGCUUGCUUAUAAGUAUUUUUCAAAAU ..((((.((((.((((.(.(((((.(..((((.....))))..).))))).).)))).))))))))............((....(((((....))))).....))..... ( -28.60) >DroYak_CAF1 62065 90 + 1 UCAAAGCACAGAGUUACCAUGCUUCCCUACGGUAUUUCAGCCACUGUGCAAGGUCACACUGUCUUG-------------------AAUGCUU-CAAGUAUUUGUAAAGUU ...(((.((((.((.(((.(((........(((......))).....))).))).)).)))))))(-------------------(((((..-...))))))........ ( -19.12) >consensus UAGAAGCACAGAGUGACCAUGCACCGCAACGGUAUUUCCGUCACUGUGCAAGGUCACACUGUCUUCACAAUCAA_AUCG______C_UGCUU_UAAGUAUUUUUCAAUAU ..((((.((((.((((((.(((((.(..((((.....))))..).))))).)))))).))))))))............................................ (-22.21 = -23.90 + 1.69)



| Location | 1,583,447 – 1,583,537 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 71.01 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -23.41 |
| Energy contribution | -26.73 |
| Covariance contribution | 3.31 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.75 |
| SVM decision value | 4.87 |
| SVM RNA-class probability | 0.999957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1583447 90 - 22407834 AAAGUGAAAAAU-------------------CGAG-UUAUUUGUGAAGACAGUGUUACCUUGCACAGUGACGGAAAUGCCGUUGCGGUGCAUGGUCACUCUGUGCUUCUA ............-------------------....-........((((((((.((.(((.(((((.(..((((.....))))..).))))).))).)).)))).)))).. ( -33.70) >DroSec_CAF1 61945 110 - 1 AUAGUGAAAAAUACUUAUAAGCACGCGACAUCGAUUUUGAUUGUGAAUACAGUAUGACCUUGCACAGUGACGGAAAUACCGUUCCGGUGCAUGGUCACUCUGUGCUUCUA ..................((((.(((((((.......)).)))))...((((..(((((.(((((.(..((((.....))))..).))))).)))))..))))))))... ( -35.70) >DroSim_CAF1 64237 110 - 1 AUUUUGAAAAAUACUUAUAAGCAAGCGACAUCGAUUUUGAUUGUGAAAACAGUAUGACCUUGCACAGUGACGGAAAUUCCGUUGCGGUGCAUGCUCACUCUGUGCUUCUA ...................((.((((..((.((((....))))))...((((..(((.(.(((((.(..((((.....))))..).))))).).)))..)))))))))). ( -31.50) >DroYak_CAF1 62065 90 - 1 AACUUUACAAAUACUUG-AAGCAUU-------------------CAAGACAGUGUGACCUUGCACAGUGGCUGAAAUACCGUAGGGAAGCAUGGUAACUCUGUGCUUUGA ..((((((.....((((-(.....)-------------------)))).((.((((......)))).))...........))))))((((((((.....))))))))... ( -23.70) >consensus AAAGUGAAAAAUACUUA_AAGCA_G______CGAU_UUGAUUGUGAAGACAGUAUGACCUUGCACAGUGACGGAAAUACCGUUGCGGUGCAUGGUCACUCUGUGCUUCUA ............................................((((((((.((((((.(((((.(((((((.....))))))).))))).)))))).)))).)))).. (-23.41 = -26.73 + 3.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:38:04 2006