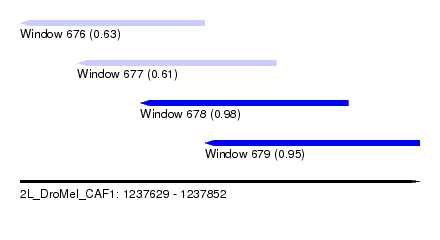
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,237,629 – 1,237,852 |
| Length | 223 |
| Max. P | 0.976263 |

| Location | 1,237,629 – 1,237,732 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.86 |
| Mean single sequence MFE | -28.27 |
| Consensus MFE | -22.09 |
| Energy contribution | -22.37 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1237629 103 - 22407834 AUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU------GCUCUUGGCAGCUG---AGUGAGCGUCGAAAGGUGAGCGAUUUUCCUCGCCCCCGAAUUUCCUCGCACAC-------- ...((((((((......))))))))..(((((..((------((.....))))..(---((.(((..(((...((((((.(.....)))))))..)))..))))))))))).-------- ( -29.30) >DroSec_CAF1 15074 107 - 1 AUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU------GCUCUUGGCAGCUG---AGUGAGCGUCGAAAGGUGAGCGAUUUUCCUCGCCCCCGAUUUUCCUCUCACACAC----AC ...((((((((......))))))))..((((...((------((.....)))).((---((.(((.((((...((((((.(.....)))))))..)))).)))..)))))))).----.. ( -27.00) >DroYak_CAF1 11132 117 - 1 AUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGCUCUUGCUCUUGGCAGCUUGGCAGUGAGCGUCGAAAGGUGAGCGAUU-UCCUCGCCCCCGGAAAA--UCACACACACACCACC ...((((((((......))))))))..((((....((((.((((.....))))...))))((((...(((...((((((.(...-.)))))))..)))....--)))).))))....... ( -28.50) >consensus AUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU______GCUCUUGGCAGCUG___AGUGAGCGUCGAAAGGUGAGCGAUUUUCCUCGCCCCCGAAUUUCCUCACACACAC_____C ...((((((((......))))))))..((((((((((((...(((......)))..))))).)))).(((...((((((.(.....)))))))..)))..........)))......... (-22.09 = -22.37 + 0.28)

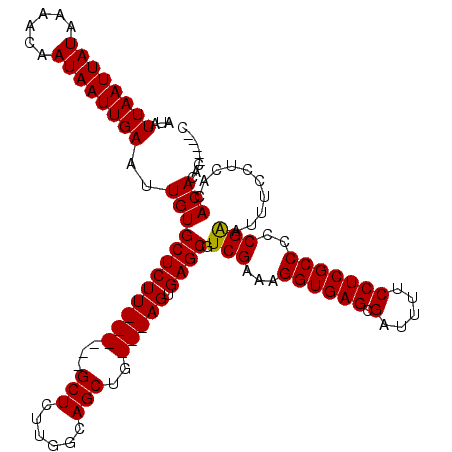

| Location | 1,237,661 – 1,237,772 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.24 |
| Mean single sequence MFE | -32.67 |
| Consensus MFE | -27.84 |
| Energy contribution | -28.09 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.612980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1237661 111 - 22407834 CAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU------GCUCUUGGCAGCUG---AGUGAGCGUCGAAAGGUGAGC ..(((..(((((((((...((.((((....((.(..(((....((((((((......)))))))).)))..)))))------))))..))))))))---.)..))).((....))..... ( -33.50) >DroSec_CAF1 15110 111 - 1 CAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU------GCUCUUGGCAGCUG---AGUGAGCGUCGAAAGGUGAGC ..(((..(((((((((...((.((((....((.(..(((....((((((((......)))))))).)))..)))))------))))..))))))))---.)..))).((....))..... ( -33.50) >DroSim_CAF1 11255 111 - 1 CAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU------GCUCUUGGCAGCUG---AGUGAACGUCGAAAGGUGAGC ..((((((((((((((...((.((((....((.(..(((....((((((((......)))))))).)))..)))))------))))..))))))))---.).)).(..(....)..)))) ( -30.90) >DroYak_CAF1 11169 120 - 1 CAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGCUCUUGCUCUUGGCAGCUUGGCAGUGAGCGUCGAAAGGUGAGC ..((((((((((....))))......((((((((((((.(((.((((((((......))))))))))).)))(((((((.((((.....))))...))))).))))))))))))).)))) ( -32.80) >consensus CAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU______GCUCUUGGCAGCUG___AGUGAGCGUCGAAAGGUGAGC ..(((...((((((((...((.((......((.(..(((....((((((((......)))))))).)))..)))........))))..))))))))...)))...(..(....)..)... (-27.84 = -28.09 + 0.25)


| Location | 1,237,696 – 1,237,812 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.88 |
| Mean single sequence MFE | -38.50 |
| Consensus MFE | -38.28 |
| Energy contribution | -38.27 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1237696 116 - 22407834 CGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU---- ((((((..(((((.......)))))..)))..(((((((((((((....)))))))))))))))).....((.(..(((....((((((((......)))))))).)))..)))..---- ( -38.10) >DroSec_CAF1 15145 116 - 1 CGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU---- ((((((..(((((.......)))))..)))..(((((((((((((....)))))))))))))))).....((.(..(((....((((((((......)))))))).)))..)))..---- ( -38.10) >DroSim_CAF1 11290 116 - 1 CGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU---- ((((((..(((((.......)))))..)))..(((((((((((((....)))))))))))))))).....((.(..(((....((((((((......)))))))).)))..)))..---- ( -38.10) >DroYak_CAF1 11209 120 - 1 CGCCCAUAAGUGCGUGGGAGGCGUUGGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUUGCUC .((.(....).))(..((((((((..(.....(((((((((((((....)))))))))))))......)..))))((((....((((((((......)))))))).)))).))))..).. ( -39.70) >consensus CGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAAAUAUUAAUUAUAAAACAAUAAUUGAAUUGUGCUCUU____ (((((((....((((.....))))..))))..(((((((((((((....)))))))))))))))).....((.(..(((....((((((((......)))))))).)))..)))...... (-38.28 = -38.27 + -0.00)

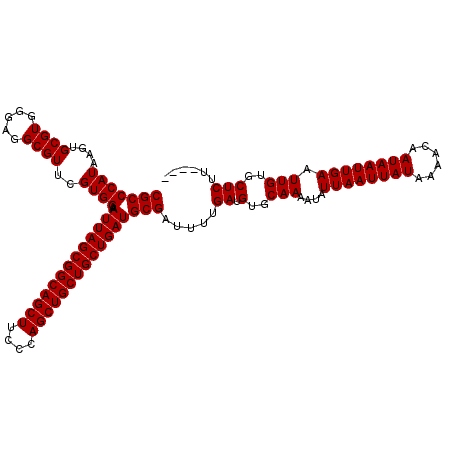
| Location | 1,237,732 – 1,237,852 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.86 |
| Mean single sequence MFE | -47.97 |
| Consensus MFE | -42.95 |
| Energy contribution | -43.83 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948075 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1237732 120 - 22407834 AACUCCCCCCCUCCAUCAACCAGUGAAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAA .............(((((((((.(((....((((.(.((((((.(....).)))))).).))))))))))..(((((((((((((....)))))))))))))......))))))...... ( -47.00) >DroSec_CAF1 15181 120 - 1 AACUCCCCCCUUCCAUCAACCGGCGAAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAA .............(((((((((.(((....((((.(.((((((.(....).)))))).).))))))))))..(((((((((((((....)))))))))))))......))))))...... ( -48.40) >DroSim_CAF1 11326 120 - 1 AGCUCCCCCUUUCCAUCAACCAGCGAAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAA .............(((((((((.(((....((((.(.((((((.(....).)))))).).))))))))))..(((((((((((((....)))))))))))))......))))))...... ( -48.80) >DroYak_CAF1 11249 119 - 1 AACUGCCCCCCUCCCCC-ACCUUUGAAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUGGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAA ...(((.........((-(((.........((((.(.((((((.(....).)))))).).)))).)))))..(((((((((((((....))))))))))))).............))).. ( -47.70) >consensus AACUCCCCCCCUCCAUCAACCAGCGAAUGUGCGUGUGCCACGCCCAUAAGUGCGUGGGAGGCGUUCGUGGAAAUUAGCGGCAGCUUCCCAGCUGCUGCUGAUGCGAUUUUGAUGUGCAAA .............(((((((((.(((....((((.(.((((((.(....).)))))).).))))))))))..(((((((((((((....)))))))))))))......))))))...... (-42.95 = -43.83 + 0.87)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:43 2006