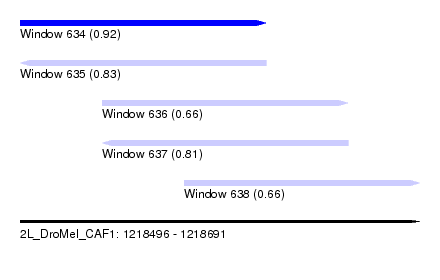
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,218,496 – 1,218,691 |
| Length | 195 |
| Max. P | 0.922748 |

| Location | 1,218,496 – 1,218,616 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.64 |
| Mean single sequence MFE | -38.00 |
| Consensus MFE | -26.69 |
| Energy contribution | -26.12 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.922748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1218496 120 + 22407834 AUUAAGUUUUGAAUCGCAUAUUGAUUAAUGCGACUAUUAUUGCGCAAUAGUAACGACGCCAGAUGGACCGAGCUGCCUUAGCUCGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAU ........(((((((........((((.((((.(.......))))).)))).....(((..((((.(((((((((...)))))))))..))))(((((.....))))).)))))))))). ( -42.00) >DroSim_CAF1 14100 120 + 1 AUUAAGUUUUGAAUCGCAUAUUGAUUAAUGCGACUAUUAUUGCGCAAUAGUAACGACGCCAGGUGGGCCGAGCUGCGUUAGCUUGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAU ........(((((((........((((.((((.(.......))))).)))).....(((..((((.(((((((((...)))))))))..))))(((((.....))))).)))))))))). ( -39.60) >DroEre_CAF1 13912 108 + 1 AUUAAGUUUUGAAUCGCAUAUUGAUUAAUGCGGCUAUGGUUGCGCAAUAGUAACGA---CAAU---------CUGCGUUAGCUUGGCAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAU ........(((((((((((((((.(((((((((...(.(((((......))))).)---....---------))))))))).....)))))).(((((.....))))).)).))))))). ( -33.90) >DroYak_CAF1 15005 111 + 1 AUUCAGUUUUGAAUCGCAUAUUGAUUAAUGCGACUGUUAUUGCGCAAUAAUAACGAUGCCAGG---------UGGCGUUAGCUUCGUAGUAUCGCCCUUUCCAAGGGCAGCGAGUUCAAU ........(((((((((.....(((...(((((..(((((((.....)))))))((((((...---------.))))))....)))))..)))(((((.....))))).)))).))))). ( -36.50) >consensus AUUAAGUUUUGAAUCGCAUAUUGAUUAAUGCGACUAUUAUUGCGCAAUAGUAACGACGCCAGG_________CUGCGUUAGCUUGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAU ........(((((((((((((((((((.((((.(.......))))).)))).......................((....))....)))))).(((((.....))))).)))).))))). (-26.69 = -26.12 + -0.56)

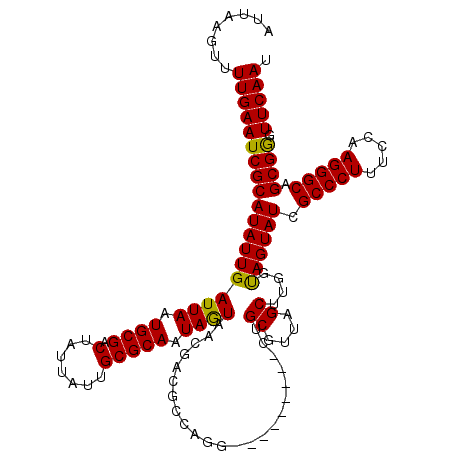
| Location | 1,218,496 – 1,218,616 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.64 |
| Mean single sequence MFE | -31.65 |
| Consensus MFE | -22.73 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1218496 120 - 22407834 AUUGAACCCGCUGCCCUUGGAAAGGGCGAUACUACCGAGCUAAGGCAGCUCGGUCCAUCUGGCGUCGUUACUAUUGCGCAAUAAUAGUCGCAUUAAUCAAUAUGCGAUUCAAAACUUAAU .(((((..(((((((((.....)))))(((...((((((((.....))))))))..))).))))......((((((.....))))))((((((........)))))))))))........ ( -38.10) >DroSim_CAF1 14100 120 - 1 AUUGAACCCGCUGCCCUUGGAAAGGGCGAUACUACCAAGCUAACGCAGCUCGGCCCACCUGGCGUCGUUACUAUUGCGCAAUAAUAGUCGCAUUAAUCAAUAUGCGAUUCAAAACUUAAU .(((((...((((((((.....)))))...........((....))))).((((((....)).))))...((((((.....))))))((((((........)))))))))))........ ( -31.30) >DroEre_CAF1 13912 108 - 1 AUUGAACCCGCUGCCCUUGGAAAGGGCGAUACUGCCAAGCUAACGCAG---------AUUG---UCGUUACUAUUGCGCAACCAUAGCCGCAUUAAUCAAUAUGCGAUUCAAAACUUAAU .(((((...((((((((.....))))).....((((((..((((((..---------...)---.)))))...))).))).....)))(((((........))))).)))))........ ( -27.50) >DroYak_CAF1 15005 111 - 1 AUUGAACUCGCUGCCCUUGGAAAGGGCGAUACUACGAAGCUAACGCCA---------CCUGGCAUCGUUAUUAUUGCGCAAUAACAGUCGCAUUAAUCAAUAUGCGAUUCAAAACUGAAU .(((((.((((.(((((.....)))))(((...((((.((((......---------..)))).))))...((.(((((.......).)))).))))).....)))))))))........ ( -29.70) >consensus AUUGAACCCGCUGCCCUUGGAAAGGGCGAUACUACCAAGCUAACGCAG_________CCUGGCGUCGUUACUAUUGCGCAAUAAUAGUCGCAUUAAUCAAUAUGCGAUUCAAAACUUAAU .((((...(((((((((.....))))))..........((....)).............................)))........(((((((........)))))))))))........ (-22.73 = -22.98 + 0.25)

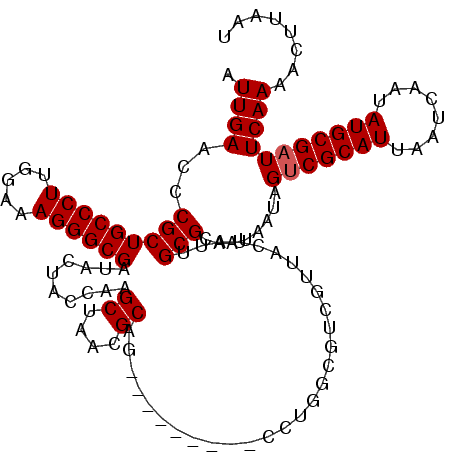
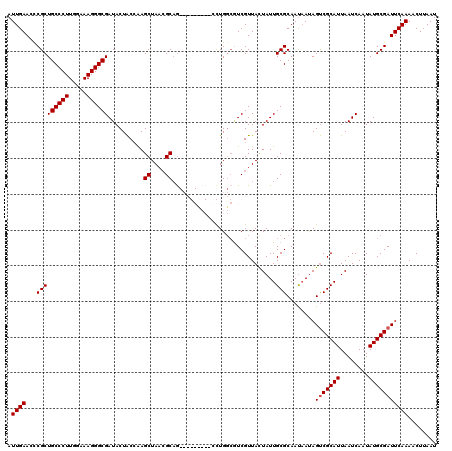
| Location | 1,218,536 – 1,218,656 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.71 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -26.35 |
| Energy contribution | -26.72 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.658422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1218536 120 + 22407834 UGCGCAAUAGUAACGACGCCAGAUGGACCGAGCUGCCUUAGCUCGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCAUGAACCGCCGAACUAUUACAAGUGGGCAAA (((.((...((((...(((..((((.(((((((((...)))))))))..))))(((((.....))))).)))((((((............)))))).........))))...)).))).. ( -43.90) >DroSim_CAF1 14140 119 + 1 UGCGCAAUAGUAACGACGCCAGGUGGGCCGAGCUGCGUUAGCUUGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCGUGAACCGCCGAACUAUUACAAGUG-GCAAA (((......))).....(((.((((((((((((((...)))))))))....(((((((.....))))).(((((((....))))).))...)).)))))..(((......))))-))... ( -43.40) >DroEre_CAF1 13952 107 + 1 UGCGCAAUAGUAACGA---CAAU---------CUGCGUUAGCUUGGCAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCGUGAACCGCCGAACUAUUACAAGUG-GCAAA (((.((...((((((.---(...---------..))))(((.(((((.((.(((((((.....))))).(((((((....))))).))...)))).))))).))).)))...))-))).. ( -31.80) >DroYak_CAF1 15045 110 + 1 UGCGCAAUAAUAACGAUGCCAGG---------UGGCGUUAGCUUCGUAGUAUCGCCCUUUCCAAGGGCAGCGAGUUCAAUAACCCAGCCAUGAACCGCCGAACUAUUACAAGUG-GCAAA (((...........((((((...---------.)))))).((((.(((((((((((((.....))))).(((.(((((............)))))))).))..)))))))))).-))).. ( -33.80) >consensus UGCGCAAUAGUAACGACGCCAGG_________CUGCGUUAGCUUGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCAUGAACCGCCGAACUAUUACAAGUG_GCAAA (((.((...((((.....................((....))(((((.((.(((((((.....))))).(((((((....))))).))...)))).)))))....))))...)).))).. (-26.35 = -26.72 + 0.37)

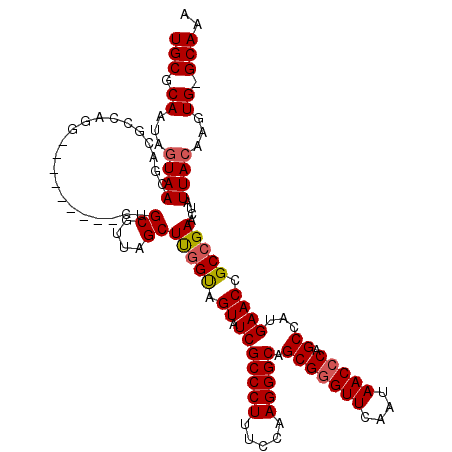
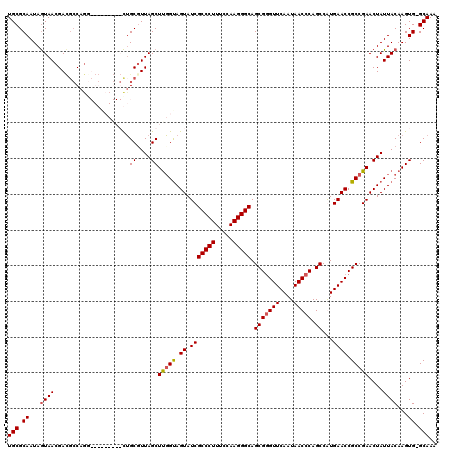
| Location | 1,218,536 – 1,218,656 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.71 |
| Mean single sequence MFE | -41.15 |
| Consensus MFE | -32.58 |
| Energy contribution | -32.70 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.811532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1218536 120 - 22407834 UUUGCCCACUUGUAAUAGUUCGGCGGUUCAUGGCUGGGUUAUUGAACCCGCUGCCCUUGGAAAGGGCGAUACUACCGAGCUAAGGCAGCUCGGUCCAUCUGGCGUCGUUACUAUUGCGCA ...........((((((((.(((((.(..((((..(((((....)))))..((((((.....)))))).....((((((((.....))))))))))))..).)))))..))))))))... ( -48.10) >DroSim_CAF1 14140 119 - 1 UUUGC-CACUUGUAAUAGUUCGGCGGUUCACGGCUGGGUUAUUGAACCCGCUGCCCUUGGAAAGGGCGAUACUACCAAGCUAACGCAGCUCGGCCCACCUGGCGUCGUUACUAUUGCGCA .....-.....((((((((.(((((.(....(((((((((....)))))((((((((.....)))))...........((....)))))..)))).....).)))))..))))))))... ( -43.30) >DroEre_CAF1 13952 107 - 1 UUUGC-CACUUGUAAUAGUUCGGCGGUUCACGGCUGGGUUAUUGAACCCGCUGCCCUUGGAAAGGGCGAUACUGCCAAGCUAACGCAG---------AUUG---UCGUUACUAUUGCGCA .....-.....((((((((.((((((((...(((.(((((....)))))..((((((.....)))))).....)))..((....)).)---------))))---)))..))))))))... ( -40.50) >DroYak_CAF1 15045 110 - 1 UUUGC-CACUUGUAAUAGUUCGGCGGUUCAUGGCUGGGUUAUUGAACUCGCUGCCCUUGGAAAGGGCGAUACUACGAAGCUAACGCCA---------CCUGGCAUCGUUAUUAUUGCGCA ..(((-..(((.....((..((((((((((((((...)))).))))).)))))..)).....)))((((((..((((.((((......---------..)))).))))...))))))))) ( -32.70) >consensus UUUGC_CACUUGUAAUAGUUCGGCGGUUCACGGCUGGGUUAUUGAACCCGCUGCCCUUGGAAAGGGCGAUACUACCAAGCUAACGCAG_________CCUGGCGUCGUUACUAUUGCGCA ...........((((((((.(((((((((..(((.(((((....))))))))(((((.....))))))).))).....((....)).................))))..))))))))... (-32.58 = -32.70 + 0.12)

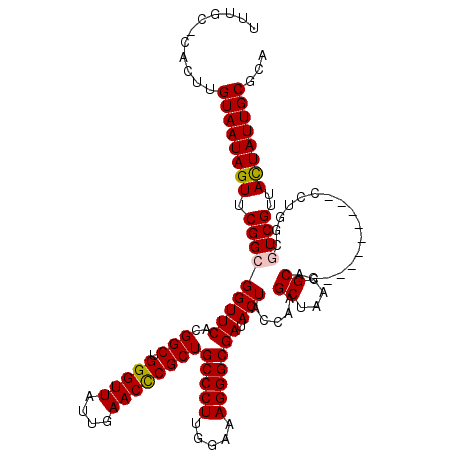
| Location | 1,218,576 – 1,218,691 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.43 |
| Mean single sequence MFE | -35.12 |
| Consensus MFE | -27.41 |
| Energy contribution | -28.10 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.661998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1218576 115 + 22407834 GCUCGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCAUGAACCGCCGAACUAUUACAAGUGGGCAAAU-----CUACUGGCCAAGGUGAGCCACUUCAAAUCACUCA ((((((..((.(((((((.....))))).(((((((....))))).))...))))..))))....((((...(((.((...-----....)).)))..))))))................ ( -34.60) >DroSim_CAF1 14180 114 + 1 GCUUGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCGUGAACCGCCGAACUAUUACAAGUG-GCAAAU-----CUACUGGCCAAGGUGAGUCACUUCAAAUCACUCA .(((((((((((((((((.....))))).(((((((....))))).))...))...(((..(((......))))-))....-----.)))).))))))((((...........))))... ( -34.00) >DroEre_CAF1 13980 119 + 1 GCUUGGCAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCGUGAACCGCCGAACUAUUACAAGUG-GCAAAUACUUGCUGCUCGCCAAGGUGAGCCACUUCAAAUCACACG ..(((((.((.(((((((.....))))).(((((((....))))).))...)))).)))))........(((((-((((....)))).((((((....)))))))))))........... ( -40.70) >DroYak_CAF1 15076 119 + 1 GCUUCGUAGUAUCGCCCUUUCCAAGGGCAGCGAGUUCAAUAACCCAGCCAUGAACCGCCGAACUAUUACAAGUG-GCAAAUAGUAGCUACUCUCCAAGGUGAGCCACUUCAAAUCACUCG .....(((((((((((((.....))))).(((.(((((............)))))))).))..))))))(((((-((.....((....))((.(....).)))))))))........... ( -31.20) >consensus GCUUGGUAGUAUCGCCCUUUCCAAGGGCAGCGGGUUCAAUAACCCAGCCAUGAACCGCCGAACUAUUACAAGUG_GCAAAU_____CUACUCGCCAAGGUGAGCCACUUCAAAUCACUCA .(((((((((((((((((.....))))).(((((((....))))).))...))...((...(((......)))..))..........)))).))))))((((...........))))... (-27.41 = -28.10 + 0.69)

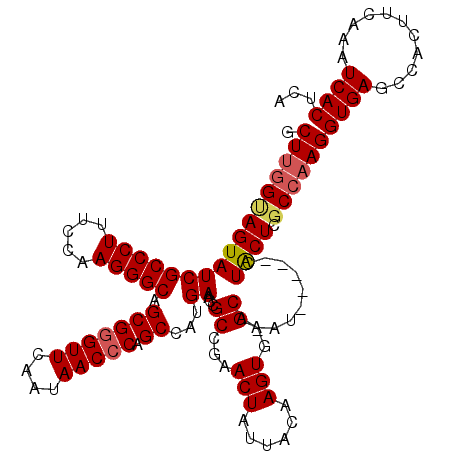
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:35:04 2006