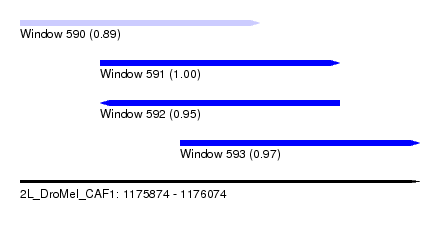
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,175,874 – 1,176,074 |
| Length | 200 |
| Max. P | 0.995246 |

| Location | 1,175,874 – 1,175,994 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.67 |
| Mean single sequence MFE | -46.18 |
| Consensus MFE | -38.48 |
| Energy contribution | -38.40 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1175874 120 + 22407834 GCAGCGGUUGCCCUCUCGGCCAUCCAGCGCAAGCAGAAGAAGAUGAGCAGUAUCGAGACCACGAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGCAAGUGAAGGCCACAUUG ((.((.((((((.(((((((((((..((....)).......)))).)).....)))))...........((((((((....))))))))...)))))).))))..(((.....))).... ( -47.60) >DroSec_CAF1 9288 120 + 1 GCAGCGGUUGCCCUCUCGGCCAUCCAGCGCAAGCAGAAGAAGAUGAACAGUAUCGAGACCACGAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGCAAGUGAAGGCCACAUUG ((.((.((((((.((((.........((....)).......((((.....))))))))...........((((((((....))))))))...)))))).))))..(((.....))).... ( -46.00) >DroEre_CAF1 10280 120 + 1 GCAGCGGUUGCCCUCUCGGCCAUCCAGCGCAAGCAGAAGAAGAUGAACAGUAUCGAGACCACAAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGGAAGUGAAGGCCACAUUG ...((.((((((.((((.........((....)).......((((.....))))))))...........((((((((....))))))))...)))))).))....(((.....))).... ( -44.20) >DroYak_CAF1 9903 120 + 1 GCAGCGGUUGCUCUCUCGGCCAUCCAGCGCAAGCAGAAGAAGAUGAGCAAUAUCGAGACCACAAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGGAAGUGAAGGCCACAUUU ((.((.(((((((((((((....)).((....))....).))).)))))))...((........))...)).))((((...((((((.....)))))).((....))...))))...... ( -46.80) >DroAna_CAF1 9676 120 + 1 GCUGCCGUGGCUCUAUCGGCUAUCCAACGCAAGCAGAAGAAGAUGAGCAACAUCGAGACCACGAUUACAGCAGCGGCAGCAGCUGCCGCCCAAGCAGCUGCUGAGGUGAAGGCUGCCUUU (((((((((((((..((.(((..........))).))....((((.....))))))).)))))......)))))..((((((((((.......))))))))))((((.......)))).. ( -46.30) >consensus GCAGCGGUUGCCCUCUCGGCCAUCCAGCGCAAGCAGAAGAAGAUGAGCAGUAUCGAGACCACGAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGGAAGUGAAGGCCACAUUG ...((.((((((.((((.........((....)).......((((.....))))))))...........((((((((....))))))))...)))))).))....(((.....))).... (-38.48 = -38.40 + -0.08)

| Location | 1,175,914 – 1,176,034 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -50.34 |
| Consensus MFE | -40.58 |
| Energy contribution | -41.22 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.81 |
| SVM decision value | 2.56 |
| SVM RNA-class probability | 0.995246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1175914 120 + 22407834 AGAUGAGCAGUAUCGAGACCACGAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGCAAGUGAAGGCCACAUUGGCUGCUCAGGUGGCCAGUGCGCCCAAGGGCAACGAUGGAA ......((.((((((......)))).)).))(((((((...((((((.....)))))).((....))...)))).(((((((..(....)..))))))).)))((..(....)..))... ( -51.10) >DroSec_CAF1 9328 120 + 1 AGAUGAACAGUAUCGAGACCACGAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGCAAGUGAAGGCCACAUUGGCUGCUCAGGUGGCCAGUGCGCCCAAGGGCAACGAUGGAA .((((.....))))....(((((......((.(.((((...((((((.....)))))).((....))...)))).)((((((..(....)..))))))))(((....)))..)).))).. ( -51.10) >DroEre_CAF1 10320 120 + 1 AGAUGAACAGUAUCGAGACCACAAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGGAAGUGAAGGCCACAUUGGCUGCUCAAGUGGUCAGUGCGCCCAAGGGCAACGAUGGAA ..........(((((.((((((.......((((((((....))))))))....(.((((((.((.(((.....))).)).)))))))..)))))).....(((....)))..)))))... ( -52.60) >DroYak_CAF1 9943 120 + 1 AGAUGAGCAAUAUCGAGACCACAAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGGAAGUGAAGGCCACAUUUGCUGCUCAGGUGGUCAGUGCGCCCAAGGGCAACGAUGGAA ..........(((((.((((((.......((((((((....))))))))....(.(((((((((.(((.....))).))))))))))..)))))).....(((....)))..)))))... ( -53.40) >DroAna_CAF1 9716 120 + 1 AGAUGAGCAACAUCGAGACCACGAUUACAGCAGCGGCAGCAGCUGCCGCCCAAGCAGCUGCUGAGGUGAAGGCUGCCUUUGCCGCCAAGAACGUAACUGCUCCAAGAGGCAACGAUGGAA ..........(((((.....(((....((((((((((((...)))))((....)).))))))).((..((((...))))..))........)))...((((......)))).)))))... ( -43.50) >consensus AGAUGAGCAGUAUCGAGACCACGAUCACAGCGGCGGCCACAGCUGCCGCACAGGCAGCGGCGGAAGUGAAGGCCACAUUGGCUGCUCAGGUGGUCAGUGCGCCCAAGGGCAACGAUGGAA ..........(((((..............((((((((....))))))))...(((....((....))....))).((((((((((....)))))))))).(((....)))..)))))... (-40.58 = -41.22 + 0.64)


| Location | 1,175,914 – 1,176,034 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -47.70 |
| Consensus MFE | -41.44 |
| Energy contribution | -41.60 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.38 |
| SVM RNA-class probability | 0.948102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1175914 120 - 22407834 UUCCAUCGUUGCCCUUGGGCGCACUGGCCACCUGAGCAGCCAAUGUGGCCUUCACUUGCGCCGCUGCCUGUGCGGCAGCUGUGGCCGCCGCUGUGAUCGUGGUCUCGAUACUGCUCAUCU ....((((..(((....))).....((((((....(((((....((((((...((..(((((((.......))))).)).)))))))).)))))....)))))).))))........... ( -47.70) >DroSec_CAF1 9328 120 - 1 UUCCAUCGUUGCCCUUGGGCGCACUGGCCACCUGAGCAGCCAAUGUGGCCUUCACUUGCGCCGCUGCCUGUGCGGCAGCUGUGGCCGCCGCUGUGAUCGUGGUCUCGAUACUGUUCAUCU ....((((..(((....))).....((((((....(((((....((((((...((..(((((((.......))))).)).)))))))).)))))....)))))).))))........... ( -47.70) >DroEre_CAF1 10320 120 - 1 UUCCAUCGUUGCCCUUGGGCGCACUGACCACUUGAGCAGCCAAUGUGGCCUUCACUUCCGCCGCUGCCUGUGCGGCAGCUGUGGCCGCCGCUGUGAUUGUGGUCUCGAUACUGUUCAUCU ....((((..(((....))).....((((((....(((((....((((((.........((.((((((.....)))))).)))))))).)))))....)))))).))))........... ( -48.00) >DroYak_CAF1 9943 120 - 1 UUCCAUCGUUGCCCUUGGGCGCACUGACCACCUGAGCAGCAAAUGUGGCCUUCACUUCCGCCGCUGCCUGUGCGGCAGCUGUGGCCGCCGCUGUGAUUGUGGUCUCGAUAUUGCUCAUCU ....((((..(((....))).....((((((....(((((....((((((.........((.((((((.....)))))).)))))))).)))))....)))))).))))........... ( -47.30) >DroAna_CAF1 9716 120 - 1 UUCCAUCGUUGCCUCUUGGAGCAGUUACGUUCUUGGCGGCAAAGGCAGCCUUCACCUCAGCAGCUGCUUGGGCGGCAGCUGCUGCCGCUGCUGUAAUCGUGGUCUCGAUGUUGCUCAUCU ..(((.(((((((.((.(((((......))))).)).))))).((((((........(((((((((((.....)))))))))))..)))))).....)))))....((((.....)))). ( -47.80) >consensus UUCCAUCGUUGCCCUUGGGCGCACUGACCACCUGAGCAGCCAAUGUGGCCUUCACUUCCGCCGCUGCCUGUGCGGCAGCUGUGGCCGCCGCUGUGAUCGUGGUCUCGAUACUGCUCAUCU ....((((.((((....))))....((((((....(((((....((((((.........((.((((((.....)))))).)))))))).)))))....)))))).))))........... (-41.44 = -41.60 + 0.16)

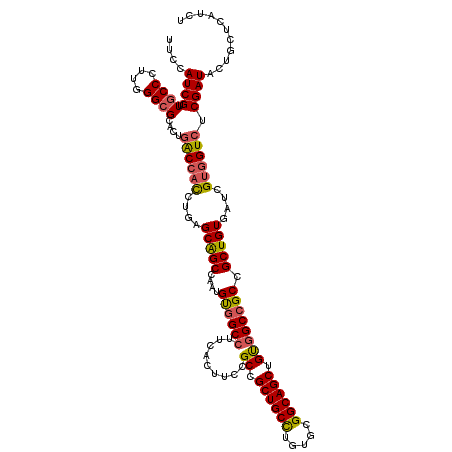

| Location | 1,175,954 – 1,176,074 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.83 |
| Mean single sequence MFE | -47.64 |
| Consensus MFE | -35.52 |
| Energy contribution | -38.68 |
| Covariance contribution | 3.16 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1175954 120 + 22407834 AGCUGCCGCACAGGCAGCGGCGCAAGUGAAGGCCACAUUGGCUGCUCAGGUGGCCAGUGCGCCCAAGGGCAACGAUGGAAAGAUAUACCGUAUGUCGCCACUCACUUGAGUAUGCUAGCA .((((((.....))))))(((((((((((.(((..(((((((..(....)..))))))).)))....(((.(((((((.........)))).))).)))..)))))))....)))).... ( -51.90) >DroSec_CAF1 9368 120 + 1 AGCUGCCGCACAGGCAGCGGCGCAAGUGAAGGCCACAUUGGCUGCUCAGGUGGCCAGUGCGCCCAAGGGCAACGAUGGAAAGAUCUACCGUAUGUCGCCACGCACUUGUGUUCUCUAGCA .((((((.....))))))((((((((((..(((..(((((((..(....)..))))))).(((....))).(((.((((....)))).))).....)))...))))))))))........ ( -55.20) >DroEre_CAF1 10360 120 + 1 AGCUGCCGCACAGGCAGCGGCGGAAGUGAAGGCCACAUUGGCUGCUCAAGUGGUCAGUGCGCCCAAGGGCAACGAUGGAAAGAUCUACCGUAAGUGACGGCAUACUAUUGUUUGCUAGCA .((((((.....)))))).((((.((......((((((((((..(....)..))))))).(((....))).....)))......)).))))..((...((((.((....)).)))).)). ( -46.50) >DroYak_CAF1 9983 120 + 1 AGCUGCCGCACAGGCAGCGGCGGAAGUGAAGGCCACAUUUGCUGCUCAGGUGGUCAGUGCGCCCAAGGGCAACGAUGGAAAGAUCUACCGUAAGUGGCCACAUACAUGUGUUUGCUAGCA .(((((.(((((.(.(((((((((.(((.....))).))))))))))..(((((((.(..(((....))).(((.((((....)))).))).).))))))).....)))))..)).))). ( -47.90) >DroAna_CAF1 9756 119 + 1 AGCUGCCGCCCAAGCAGCUGCUGAGGUGAAGGCUGCCUUUGCCGCCAAGAACGUAACUGCUCCAAGAGGCAACGAUGGAAAGGUCUACCGUAAGUAUUCG-AUAGAAGAAAAUGUUCGCA ((((((.......))))))((...((..((((...))))..)).....((((((..((..((((...(....)..)))).)).((((.((........))-.)))).....)))))))). ( -36.70) >consensus AGCUGCCGCACAGGCAGCGGCGGAAGUGAAGGCCACAUUGGCUGCUCAGGUGGUCAGUGCGCCCAAGGGCAACGAUGGAAAGAUCUACCGUAAGUCGCCACAUACUUGUGUUUGCUAGCA .((((((.....))))))((((((((((..(((((((((((((((....)))))))))..(((....))).(((.((((....)))).)))..))))))...))))))))))........ (-35.52 = -38.68 + 3.16)

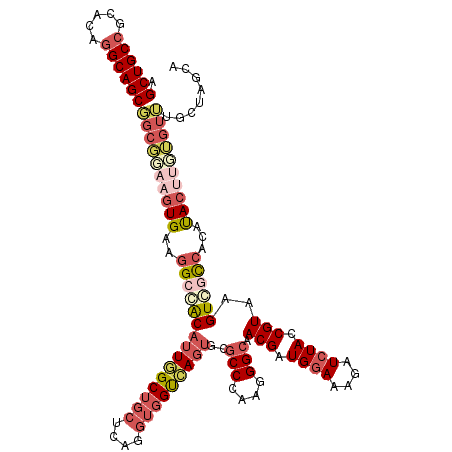
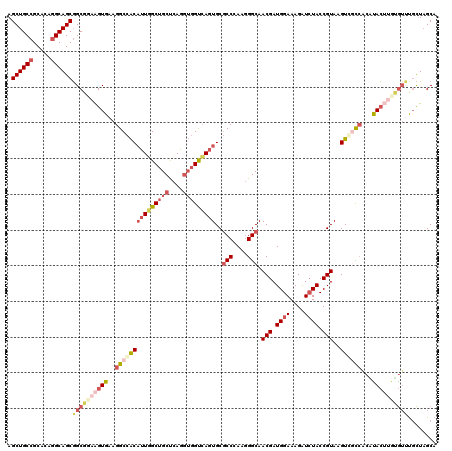
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:34:19 2006