
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
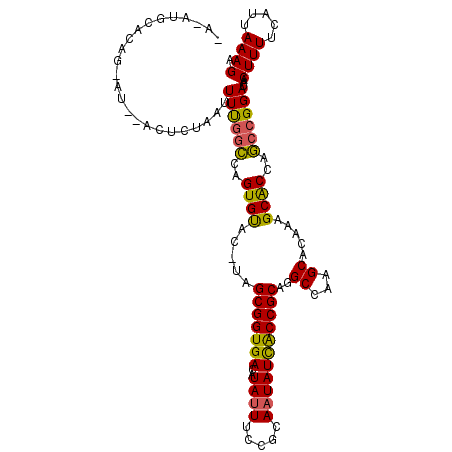
| Location | 9,222,828 – 9,222,922 |
| Length | 94 |
| Max. P | 0.777935 |

| Location | 9,222,828 – 9,222,922 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -25.18 |
| Consensus MFE | -22.60 |
| Energy contribution | -22.05 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.662988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9222828 94 + 22407834 -------------------CUUAUUUUGGCCAGUGCACUUAGCGGUGACAUAUUUCCGCAAUAUCACCGCAGGCCAAGCACAAAGCACCAGCCGGACAACUUUUCAUUAAAGA -------------------.....((((((..((((.....(((((((..((((.....)))))))))))..((...)).....))))..))))))...((((.....)))). ( -25.80) >DroSec_CAF1 52927 113 + 1 CAUAUGCACAUUAUCUACUCUAAUUUUGGCCAGUGUACUUAGCGGUGACAUAUUUCCGCAAUAUCACCGCAGGCCAAGCACAAAGCACCAGCCGGACAACUUUUCAUUAAAGA .....((..................((((((((....))..(((((((..((((.....))))))))))).))))))((.....))....)).......((((.....)))). ( -24.20) >DroSim_CAF1 54813 113 + 1 CAUAUGCAUAGUAUCUACUCUAAUUUUGGCCAGUGUACUUAGCGGUGACAUAUUUCCGCAAUAUCACCGCAGGCCAAGCACAAAGCACCAGCCGGACAACUUUUCAUUAAAGA ....(((.(((........)))..(((((((((....))..(((((((..((((.....))))))))))).)))))))......)))((....))....((((.....)))). ( -25.60) >DroEre_CAF1 59426 112 + 1 UACAUGCACCGAGUGCACUUUCGUUUUGGCCAGUGUAA-GAGCGGUGACAUAUUUCCGCAAUAUCACCGCAGGCCAAGCACAAAGCACCAGCCGGACAACUUUUCAUUAAAGA ........(((.((((((....)).((((((..(....-).(((((((..((((.....))))))))))).))))))))))...((....)))))....((((.....)))). ( -28.10) >DroYak_CAF1 58129 112 + 1 UAAAUGCACAGAAUGCACUUUAGUUUUGGCCAGUGUAA-GAGCGGUGACAUAUUUCCGCAAUAUCACCGCAGGCCAAGCACAAAGCGCCAGCCGGACAACUUUUCAUUAAAGA ....((((.....))))(((((((((((((........-..(((((((..((((.....))))))))))).(((...((.....))))).)))))).........))))))). ( -28.10) >DroAna_CAF1 54502 91 + 1 --------------------UCCUUUCUCUCAGUGCAU-UGGCGGUGACAUAUUUCCGCA-UAUUGCCGCAGGCCAAGCACAAAGCACCAAACGGACAACUUUUCAUUAAAGA --------------------............((((.(-(((((....)........((.-.......))..)))))))))......((....))....((((.....)))). ( -19.30) >consensus _A_AUGCACAG_AU__ACUCUAAUUUUGGCCAGUGUAC_UAGCGGUGACAUAUUUCCGCAAUAUCACCGCAGGCCAAGCACAAAGCACCAGCCGGACAACUUUUCAUUAAAGA ........................((((((..((((.....(((((((..((((.....)))))))))))..((...)).....))))..))))))...((((.....)))). (-22.60 = -22.05 + -0.55)

| Location | 9,222,828 – 9,222,922 |
|---|---|
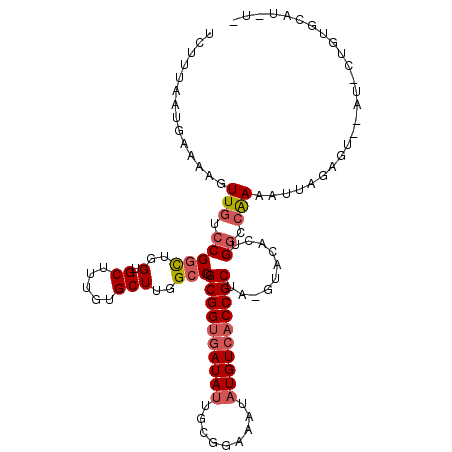
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -33.18 |
| Consensus MFE | -23.71 |
| Energy contribution | -24.43 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9222828 94 - 22407834 UCUUUAAUGAAAAGUUGUCCGGCUGGUGCUUUGUGCUUGGCCUGCGGUGAUAUUGCGGAAAUAUGUCACCGCUAAGUGCACUGGCCAAAAUAAG------------------- ....................(((..((((.....(((((((....((((((((.........)))))))))))))))))))..)))........------------------- ( -32.40) >DroSec_CAF1 52927 113 - 1 UCUUUAAUGAAAAGUUGUCCGGCUGGUGCUUUGUGCUUGGCCUGCGGUGAUAUUGCGGAAAUAUGUCACCGCUAAGUACACUGGCCAAAAUUAGAGUAGAUAAUGUGCAUAUG ......(((....((((((.......(((((((...((((((.((((((((((.........))))))))))..((....))))))))...)))))))))))))...)))... ( -33.71) >DroSim_CAF1 54813 113 - 1 UCUUUAAUGAAAAGUUGUCCGGCUGGUGCUUUGUGCUUGGCCUGCGGUGAUAUUGCGGAAAUAUGUCACCGCUAAGUACACUGGCCAAAAUUAGAGUAGAUACUAUGCAUAUG .....................((((((((((((...((((((.((((((((((.........))))))))))..((....))))))))...))))))....)))).))..... ( -32.30) >DroEre_CAF1 59426 112 - 1 UCUUUAAUGAAAAGUUGUCCGGCUGGUGCUUUGUGCUUGGCCUGCGGUGAUAUUGCGGAAAUAUGUCACCGCUC-UUACACUGGCCAAAACGAAAGUGCACUCGGUGCAUGUA ............((((....)))).(..((..((((((((((.((((((((((.........))))))))))..-.......)))))).((....))))))..))..)..... ( -38.30) >DroYak_CAF1 58129 112 - 1 UCUUUAAUGAAAAGUUGUCCGGCUGGCGCUUUGUGCUUGGCCUGCGGUGAUAUUGCGGAAAUAUGUCACCGCUC-UUACACUGGCCAAAACUAAAGUGCAUUCUGUGCAUUUA ...............((..(((...(((((((((..((((((.((((((((((.........))))))))))..-.......)))))).)).)))))))...)))..)).... ( -39.10) >DroAna_CAF1 54502 91 - 1 UCUUUAAUGAAAAGUUGUCCGUUUGGUGCUUUGUGCUUGGCCUGCGGCAAUA-UGCGGAAAUAUGUCACCGCCA-AUGCACUGAGAGAAAGGA-------------------- (((((.....(((((...((....)).)))))(((((((((..(.((((.((-(......))))))).).))))-).))))..))))).....-------------------- ( -23.30) >consensus UCUUUAAUGAAAAGUUGUCCGGCUGGUGCUUUGUGCUUGGCCUGCGGUGAUAUUGCGGAAAUAUGUCACCGCUA_GUACACUGGCCAAAAUUAGAGU__AU_CUGUGCAU_U_ ..............(((.(((((..(.((.....)))..))).((((((((((.........))))))))))..........)).)))......................... (-23.71 = -24.43 + 0.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:47:18 2006