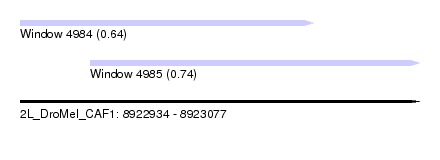
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 8,922,934 – 8,923,077 |
| Length | 143 |
| Max. P | 0.741319 |

| Location | 8,922,934 – 8,923,039 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -30.26 |
| Consensus MFE | -21.16 |
| Energy contribution | -21.02 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.636032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
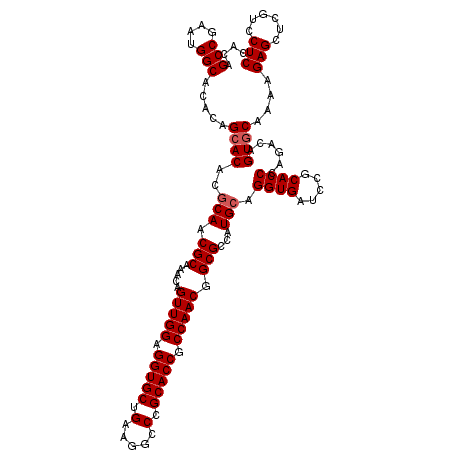
>2L_DroMel_CAF1 8922934 105 + 22407834 ---------------CAGAUAUCCGACUCCCAAAACAUCUCACAGCCAAAUGGCACACAGCACACGCAACGCAAACAGUUGGAGGUGCUGAAGGCCCGCACCGCCAACGGCGCCAUGCAG ---------------......(((((((................(((....))).....((....)).........)))))))(((((((..(((.......)))..)))))))...... ( -31.80) >DroVir_CAF1 102191 114 + 1 UCUUGCCCGAUUUUGCAGAUAUCAGAUUCCCAAAAUAUCUCACAGCCGAAUGGCACACAGCACACACAGCGCAAACAAAUGGAGGUGCUG------CGCACCGCCAAUGGCGCCAUGCAG ...(((.......(((((((((............))))))....(((....))).....)))......((((.......(((.(((((..------.))))).)))...))))...))). ( -27.60) >DroGri_CAF1 101536 111 + 1 U---GAUCAACAUUGCAGAUAUCGGAUUCACAAAAUAUCUCACAGCCGAAUGGCACACAGCACAUACAGCGCAAACAAAUGGAGGUACUA------CGAAGCGCCAAUGGUGCCAUGCAG .---.........(((.....((((....................))))(((((((.((((.(.....).))...........((..((.------...))..))..)))))))))))). ( -23.85) >DroWil_CAF1 196850 105 + 1 ---------------CAGAUAUCCGAUUCCCAAAACAUCUCACAGCCAAAUGGCACACAGCACACACAGCGCAAACAGUUGGAGGUGCUGCAGGCACGCACCGCCAACGGCGCCAUGCAA ---------------.............................(((....))).....(((......((((.....(((((.(((((((....)).))))).))))).))))..))).. ( -31.40) >DroYak_CAF1 137404 105 + 1 ---------------CAGAUAUCCGACUCCCAAAACAUCUCACAGCCGAAUGGCACACAGCACACGCAGCGCAAACAGUUGGAGGUGCUGAAGGCCCGCACCGCCAACGGCGCCAUGCAG ---------------......(((((((................(((....))).....((.(.....).))....)))))))(((((((..(((.......)))..)))))))...... ( -31.50) >DroMoj_CAF1 164964 114 + 1 UCUCGUGCGAUAUUGCAGAUAUCCGAUUCCCAAAAUAUCUCACAGCCGAAUGGCACACAGCACACACAGCGCAAGCAAAUGGAGGUGCUG------CGCACCGCCAAUGGCGCCAUGCAG ....(((.((((((...((........))....)))))).))).((...(((((.(.((((((.((..((....))...))...))))))------.)....(((...)))))))))).. ( -35.40) >consensus _______________CAGAUAUCCGAUUCCCAAAACAUCUCACAGCCGAAUGGCACACAGCACACACAGCGCAAACAAAUGGAGGUGCUG______CGCACCGCCAACGGCGCCAUGCAG ............................................(((....))).....(((......((((.....(.(((.(((((.........))))).))).).))))..))).. (-21.16 = -21.02 + -0.14)



| Location | 8,922,959 – 8,923,077 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 96.84 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -37.10 |
| Energy contribution | -37.43 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.741319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 8922959 118 + 22407834 CACAGCCAAAUGGCACACAGCACACGCAACGCAAACAGUUGGAGGUGCUGAAGGCCCGCACCGCCAACGGCGCCAUGCAGGUGAUCCGCACCCAGACAGUGCAAAAGAGUUCGUCCUC ....(((....))).....((((..(((.(((.....(((((.(((((.(.....).))))).))))).)))...))).((((.....))))......))))....(((......))) ( -39.50) >DroSec_CAF1 133141 118 + 1 CACAGCCGAAUGGCACACAGCACACGCAACGCAAACAGUUGGAGGUGCUGAAGGCCCGCACCGCCAACGGCGCCAUGCAGGUGAUCCGCACCCAGACAGUCCAAAAGAGCUCGUCCUC ....((((..((((...((((((...((((.......))))...))))))..((......)))))).))))........((((.....))))..(((((..(....)..)).)))... ( -37.20) >DroSim_CAF1 132183 118 + 1 CACAGCCGAAUGGCACACAGCACACGCAACGCAAACAGUUGGAGGUGCUGAAGGCCCGCACCGCCAACGGCGCCAUGCAGGUGAUCCGCACCCAGACCGUGCAAAAGAGCUCGUCCUC ....(((....))).....((((..(((.(((.....(((((.(((((.(.....).))))).))))).)))...))).((((.....))))......))))................ ( -38.60) >DroEre_CAF1 141028 118 + 1 CACAGCCGAAUGGCACACAGCACACGCAGCGCAAACAGUUGGAGGUGCUGAAGGCCCGCACCGCCAACGGCGCCAUGCAGGUGAUCCGCACCCAGACCGUGCAAAAGAGCUCGUCCUC ....(((....))).....((((..(((((((.....(((((.(((((.(.....).))))).))))).))))..))).((((.....))))......))))................ ( -42.10) >DroYak_CAF1 137429 118 + 1 CACAGCCGAAUGGCACACAGCACACGCAGCGCAAACAGUUGGAGGUGCUGAAGGCCCGCACCGCCAACGGCGCCAUGCAGGUGAUCCGCACCCAGACGGUGCAAAAGAGCUCGUCCUC ....(((....)))..(((((.((((((((((.....(((((.(((((.(.....).))))).))))).))))..)))..)))....(((((.....)))))......))).)).... ( -43.40) >DroAna_CAF1 135493 118 + 1 CACAGCCGAACGGCACACAGCACACACAGCGCAAACAGUUGGAGGUGCUGAAGGCCCGCACCGCCAACGGCGCCAUGCAGGUGAUCCGCACCCAGACGGUGCAGAAGAGCUCCUCCUC ....(((....)))....(((.(((.((((((.....(((((.(((((.(.....).))))).))))).))))..))...))).((.(((((.....))))).))...)))....... ( -42.20) >consensus CACAGCCGAAUGGCACACAGCACACGCAACGCAAACAGUUGGAGGUGCUGAAGGCCCGCACCGCCAACGGCGCCAUGCAGGUGAUCCGCACCCAGACAGUGCAAAAGAGCUCGUCCUC ....(((....))).....((((..(((.(((.....(((((.(((((.(.....).))))).))))).)))...))).((((.....))))......))))....(((......))) (-37.10 = -37.43 + 0.33)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:45:11 2006