| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 876,694 – 876,791 |
| Length | 97 |
| Max. P | 0.761910 |

| Location | 876,694 – 876,791 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -17.58 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
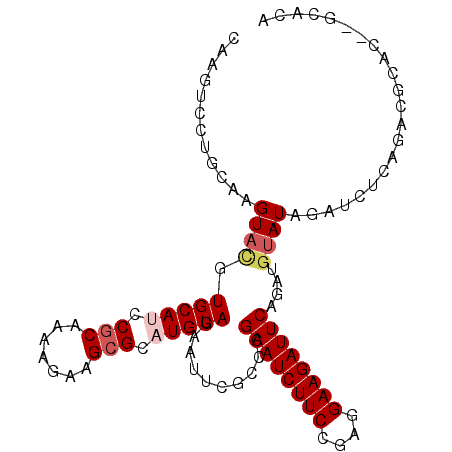
>2L_DroMel_CAF1 876694 97 + 22407834 CAAGAACUGCAAGUAUGUGCAUCCGCAGAAGAAGCGCUUGUAGGACCUACCCGAAUCUUCUGCGGAAGAUUCAGCUGCAUAGAUCUCAGACGCGC----AA- ...............(((((.(((((((((((..((...((((...)))).))..)))))))))))((((..(......)..)))).....))))----).- ( -31.60) >DroVir_CAF1 4843 100 + 1 CAAGUCGUGCAAGUACGUGCAUCCGCAAAAGAAGAGAAUGUAGGACUCAUCCGAAUCUUCCGAGGAAGAUUCGAAAGUAUAGAUCUCACAGAGCC--UCACA ...((.(((((......)))))..))............((((((.(((...(((((((((....)))))))))........(....)...)))))--).))) ( -24.40) >DroPse_CAF1 4837 102 + 1 CAAGUCUUGCAAGUACGUGCACCCGCAAAAUAAGCGCAUGUAGAAUUUGCCCGAAUCUUCCCAGGAAGAUUCAGUUGUAUAGAUCUCAGACGCAGACGCACA ...((((.((((((.(.((((..(((.......)))..))))).))))))..((((((((....))))))))...............))))((....))... ( -28.50) >DroMoj_CAF1 4927 100 + 1 CAAGUCCUGCAAGUACGUGCAUCCGCAGAAGAAGCGUAUGUAGGAUUCGGCCGAAUCUUCCGAGGAAGAUUCGCAUAUAUAGAUCUCACAGAGCG--CCACA ..(((((((((..(((((...((....))....)))))))))))))).((((((((((((....)))))))))...........(((...))).)--))... ( -31.80) >DroAna_CAF1 4970 102 + 1 CAAGAACUGCAAGUACGUGCAUCCGCAGAAGAAGCGCAUGUAGAAUUCGGCCGAAUCUUCUGCGGAAGAUUCAACUGUAUAGAUCUCAGACGAUCGCUCAAA ........((..((((((.(.(((((((((((..((..((.......))..))..))))))))))).).....)).)))).((((......))))))..... ( -29.00) >DroPer_CAF1 4835 102 + 1 CAAGUCUUGCAAGUACGUGCAUCCGCAAAAUAAGCGCAUGUAGAAUUUGCCCGAAUCUUCCCAGGAAGAUUCAGUUGUAUAGAUCUCAGACGCAGACGCACA ...((((.((((((.(.(((((.(((.......))).)))))).))))))..((((((((....))))))))...............))))((....))... ( -30.10) >consensus CAAGUCCUGCAAGUACGUGCAUCCGCAAAAGAAGCGCAUGUAGAAUUCGCCCGAAUCUUCCGAGGAAGAUUCAGAUGUAUAGAUCUCAGACGCAC__GCACA ............((((.(((((.(((.......))).)))))..........((((((((....))))))))....))))...................... (-17.58 = -18.30 + 0.72)

| Location | 876,694 – 876,791 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.25 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -18.79 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.736705 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
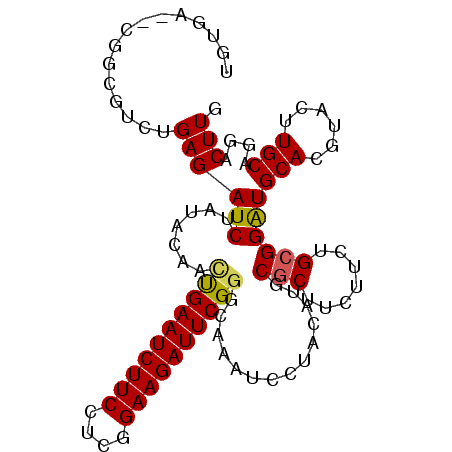
>2L_DroMel_CAF1 876694 97 - 22407834 -UU----GCGCGUCUGAGAUCUAUGCAGCUGAAUCUUCCGCAGAAGAUUCGGGUAGGUCCUACAAGCGCUUCUUCUGCGGAUGCACAUACUUGCAGUUCUUG -..----((((((..(....).)))).)).((((..(((((((((((..((.((((...))))...))..)))))))))))((((......))))))))... ( -31.70) >DroVir_CAF1 4843 100 - 1 UGUGA--GGCUCUGUGAGAUCUAUACUUUCGAAUCUUCCUCGGAAGAUUCGGAUGAGUCCUACAUUCUCUUCUUUUGCGGAUGCACGUACUUGCACGACUUG ((..(--(((((((..(((........(((((((((((....))))))))))).(((.........)))...)))..)))).)).....))..))....... ( -28.90) >DroPse_CAF1 4837 102 - 1 UGUGCGUCUGCGUCUGAGAUCUAUACAACUGAAUCUUCCUGGGAAGAUUCGGGCAAAUUCUACAUGCGCUUAUUUUGCGGGUGCACGUACUUGCAAGACUUG .....((((((((.(((((.........((((((((((....))))))))))......))).)).))))......((((((((....))))))))))))... ( -35.86) >DroMoj_CAF1 4927 100 - 1 UGUGG--CGCUCUGUGAGAUCUAUAUAUGCGAAUCUUCCUCGGAAGAUUCGGCCGAAUCCUACAUACGCUUCUUCUGCGGAUGCACGUACUUGCAGGACUUG ..(((--(.(((...)))...........(((((((((....)))))))))))))..((((.....(((.......)))...(((......))))))).... ( -28.90) >DroAna_CAF1 4970 102 - 1 UUUGAGCGAUCGUCUGAGAUCUAUACAGUUGAAUCUUCCGCAGAAGAUUCGGCCGAAUUCUACAUGCGCUUCUUCUGCGGAUGCACGUACUUGCAGUUCUUG ...((((((((......))))...............(((((((((((..((...............))..)))))))))))((((......))))))))... ( -30.66) >DroPer_CAF1 4835 102 - 1 UGUGCGUCUGCGUCUGAGAUCUAUACAACUGAAUCUUCCUGGGAAGAUUCGGGCAAAUUCUACAUGCGCUUAUUUUGCGGAUGCACGUACUUGCAAGACUUG .(((((((((((..((((..........((((((((((....))))))))))(((.........))).))))...)))))))))))................ ( -36.90) >consensus UGUGA__CGGCGUCUGAGAUCUAUACAACUGAAUCUUCCUCGGAAGAUUCGGGCAAAUCCUACAUGCGCUUCUUCUGCGGAUGCACGUACUUGCAGGACUUG ...............((((((.......((((((((((....))))))))))..............(((.......))))))(((......)))....))). (-18.79 = -18.60 + -0.19)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:32:34 2006