
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 6,135,827 – 6,135,978 |
| Length | 151 |
| Max. P | 0.976351 |

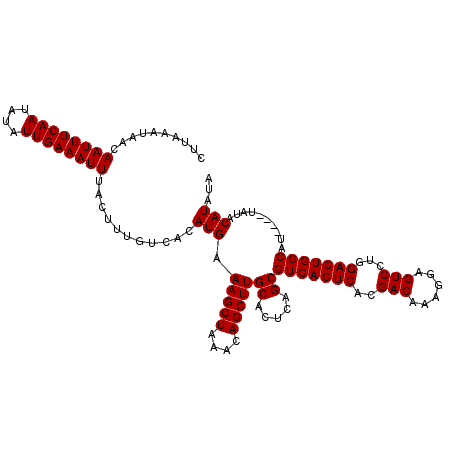
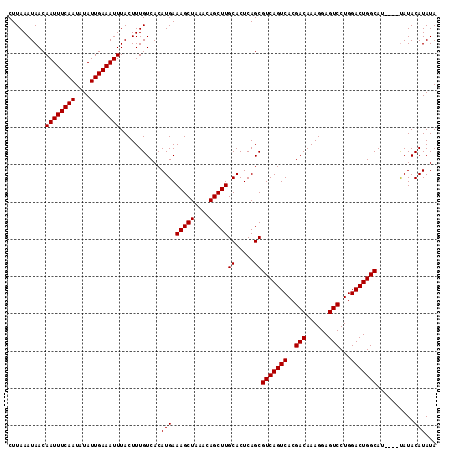
| Location | 6,135,827 – 6,135,938 |
|---|---|
| Length | 111 |
| Sequences | 3 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 96.48 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -23.10 |
| Energy contribution | -23.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.02 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.22 |
| SVM RNA-class probability | 0.932413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6135827 111 - 22407834 CUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGACAAAGGAGUCCUGGACUGGCAA----UAUACAUAUA ..........((((((((....))))))))...........(((.(((((....)))))((.....))(((((((..(((......)))...)))))))..----....)))... ( -23.10) >DroSim_CAF1 4513 111 - 1 CUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGACAAAGGAGUCCUGGACUGGCAU----UAUACAUAUA ..........((((((((....))))))))...........(((.(((((....)))))((.....))(((((((..(((......)))...)))))))..----....)))... ( -23.10) >DroEre_CAF1 4780 115 - 1 CUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGACAAAGGAGUCCUGGACUGGCAUCGCACAUACAUAUA ..........((((((((....)))))))).....((((...((.(((((....))))).))......(((((((..(((......)))...)))))))...).)))........ ( -23.20) >consensus CUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGACAAAGGAGUCCUGGACUGGCAU____UAUACAUAUA ..........((((((((....))))))))...........(((.(((((....)))))((.....))(((((((..(((......)))...)))))))..........)))... (-23.10 = -23.10 + -0.00)

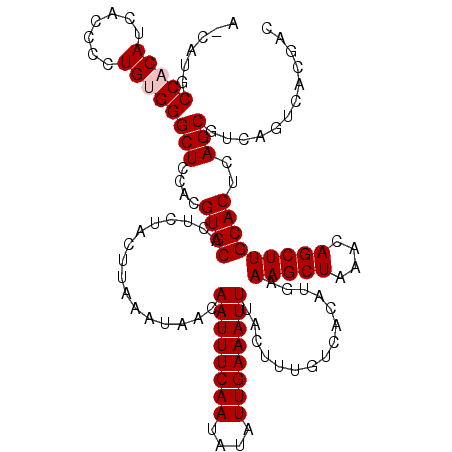
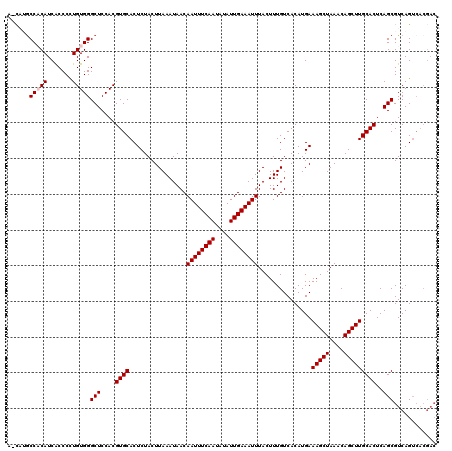
| Location | 6,135,858 – 6,135,978 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.30 |
| Mean single sequence MFE | -22.82 |
| Consensus MFE | -19.66 |
| Energy contribution | -20.06 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.976351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 6135858 120 - 22407834 ACCAUGCCACAUCACCCCUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGAC ......(((((.......)))))(((....((((................((((((((....))))))))...............(((((....)))))))))..)))(((......))) ( -22.40) >DroSec_CAF1 4480 113 - 1 A-CAUGCCCCAUCACCCCUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGA------ .-.(((((((((.......)))))......((((................((((((((....))))))))...............(((((....)))))))))...))))....------ ( -21.20) >DroSim_CAF1 4544 119 - 1 A-CAUGCCCCAUCACCCCUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGAC .-...((((((.......)).))))....((((.((..............((((((((....))))))))............((((((((....)))))((.....)).))))))))).. ( -22.60) >DroEre_CAF1 4815 119 - 1 A-CCAACCACAUCACCCUUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGAC .-....(((((.......)))))(((....((((................((((((((....))))))))...............(((((....)))))))))..)))(((......))) ( -23.50) >DroYak_CAF1 4782 119 - 1 A-UCAGCCACAUCGCCCUUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGAC .-...((......((((....)))).....((((................((((((((....))))))))...............(((((....)))))))))...))(((......))) ( -24.40) >consensus A_CAUGCCACAUCACCCCUGUGGGCUCCACGUGCACUCUACUUAAAUAACAAUUUCAAUAUAUUGAAAUUUACUUUGUCACAUGAAAGCUAAACAGCUUGCACUCAGCGUCAGUCACGAC ......(((((.......)))))(((....((((................((((((((....))))))))...............(((((....)))))))))..)))............ (-19.66 = -20.06 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:23:08 2006