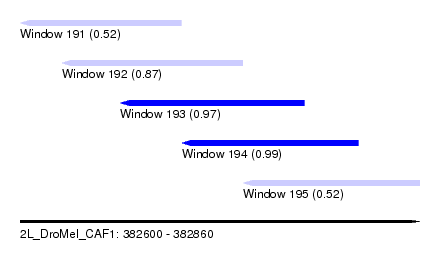
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
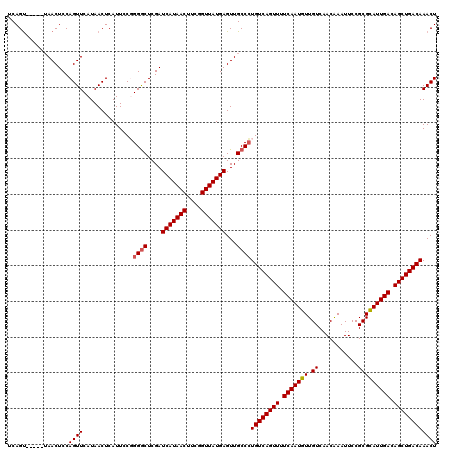
| Location | 382,600 – 382,860 |
| Length | 260 |
| Max. P | 0.992108 |

| Location | 382,600 – 382,705 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -23.40 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.40 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 382600 105 - 22407834 UAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGUUGCGAUUGCCAC--------CGACUUCAUUA-----CAAGACCAG--CACUGAU .....(((((((.((((..........((((((((......))))))))(((.(((((...))))).))).))))...--------.(....)....-----...)))).)--))..... ( -22.60) >DroSec_CAF1 50238 105 - 1 UAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGCUGCGAUUGCCCC--------CGACUUCAUUA-----CAAGACCAG--CACUGAU .....(((((((.((((..........((((((((......))))))))(((..((((...))))..))).))))...--------.(....)....-----...)))).)--))..... ( -21.20) >DroSim_CAF1 50928 105 - 1 UAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGUUGCGAUUGCCCC--------CGACUUCAUUA-----CAAGACCAG--CACUGAU .....(((((((.((((..........((((((((......))))))))(((.(((((...))))).))).))))...--------.(....)....-----...)))).)--))..... ( -22.60) >DroYak_CAF1 51527 120 - 1 UAAUUUGCGGACAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGUUGCAAUUUCCCCCCACCCCGCGACUUCAUUACAGUACAAGACCAGCGCACUGAU ....(((((((((((.((((....)))).)))))..(((.((((((((((((.....)))..))))))))).)))........))))))........((((.(.........).)))).. ( -27.20) >consensus UAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGUUGCGAUUGCCCC________CGACUUCAUUA_____CAAGACCAG__CACUGAU ((((((((.....))))))))......((((((((......))))))))(((.(((((...))))).)))......................................(((....))).. (-18.34 = -18.40 + 0.06)

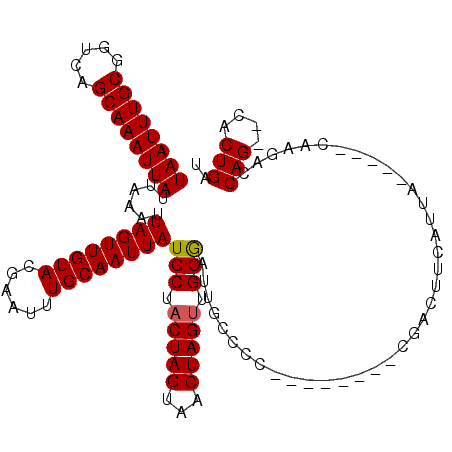
| Location | 382,627 – 382,745 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.22 |
| Mean single sequence MFE | -28.25 |
| Consensus MFE | -25.64 |
| Energy contribution | -25.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 382627 118 - 22407834 AUGUUGUCAACAAAUUCCGCGUAUUGACAGCUGACAAACUUAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGUUGCGAUUGCCAC-- ..((.(((..........(((..(((((((((((......((((((((.....)))))))).....)))))))).)))...)))......((.(((((...))))).))))).))...-- ( -29.10) >DroSec_CAF1 50265 118 - 1 AUGUUGUCAACAAAUUCCGCGCAUUGACAGCUGACAAACUUAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGCUGCGAUUGCCCC-- ..((.(((............((((((((((((((......((((((((.....)))))))).....)))))))).)))...)))......((..((((...))))..))))).))...-- ( -27.20) >DroSim_CAF1 50955 118 - 1 AUGUUGUCAACAAAUUCCGCGCAUUGACAGCUGACAAACUUAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGUUGCGAUUGCCCC-- ..((.(((............((((((((((((((......((((((((.....)))))))).....)))))))).)))...)))......((.(((((...))))).))))).))...-- ( -28.60) >DroYak_CAF1 51567 120 - 1 AUGUUGUCAACGAAUUCCGCGCAUUGACAGCUGACAAACUUAAUUUGCGGACAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGUUGCAAUUUCCCCCC ..((((((((((.......))..))))))))..((((.((((((((((.....)))))))).......))))))..(((.((((((((((((.....)))..))))))))).)))..... ( -28.11) >consensus AUGUUGUCAACAAAUUCCGCGCAUUGACAGCUGACAAACUUAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAUUUGCAAUUAUGCUACUAGUAACUAGUUGCGAUUGCCCC__ ..................(((..(((((((((((......((((((((.....)))))))).....)))))))).)))...))).....(((.(((((...))))).))).......... (-25.64 = -25.70 + 0.06)

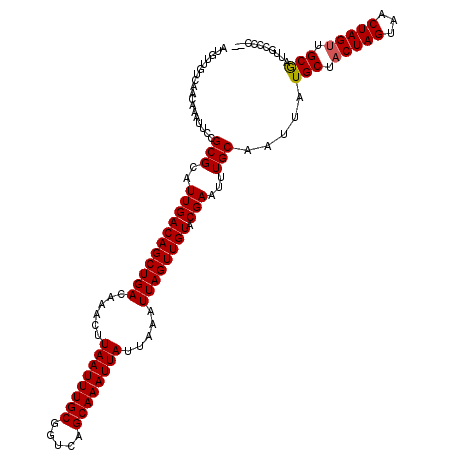
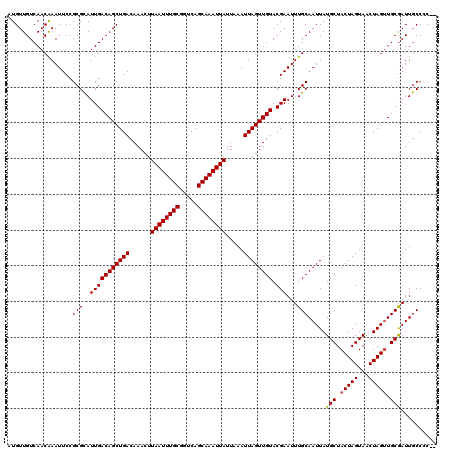
| Location | 382,665 – 382,785 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -34.25 |
| Consensus MFE | -34.11 |
| Energy contribution | -33.92 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.33 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 382665 120 - 22407834 CGAUCAUAACUUCGGUUAUGAGUUGCCAUGUCAGUUUUCAAUGUUGUCAACAAAUUCCGCGUAUUGACAGCUGACAAACUUAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAU ((.(((((((....)))))))..(((..((((((((.(((((((((....)))........)))))).))))))))....((((((((.....))))))))...........)))))... ( -31.70) >DroSec_CAF1 50303 120 - 1 CGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCAAUGUUGUCAACAAAUUCCGCGCAUUGACAGCUGACAAACUUAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAU ((.(((((((....)))))))..(((..((((((((.(((((((.((...........))))))))).))))))))....((((((((.....))))))))...........)))))... ( -35.10) >DroSim_CAF1 50993 120 - 1 CGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCAAUGUUGUCAACAAAUUCCGCGCAUUGACAGCUGACAAACUUAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAU ((.(((((((....)))))))..(((..((((((((.(((((((.((...........))))))))).))))))))....((((((((.....))))))))...........)))))... ( -35.10) >DroYak_CAF1 51607 120 - 1 CGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCAAUGUUGUCAACGAAUUCCGCGCAUUGACAGCUGACAAACUUAAUUUGCGGACAGCAAAUUAUUAAAUUAGUUGUACGAAU ((.(((((((....)))))))..(((..((((((((.(((((((.((...........))))))))).))))))))....((((((((.....))))))))...........)))))... ( -35.10) >consensus CGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCAAUGUUGUCAACAAAUUCCGCGCAUUGACAGCUGACAAACUUAAUUUGCGGUCAGCAAAUUAUUAAAUUAGUUGUACGAAU ((.(((((((....)))))))..(((..((((((((.(((((((.((...........))))))))).))))))))....((((((((.....))))))))...........)))))... (-34.11 = -33.92 + -0.19)

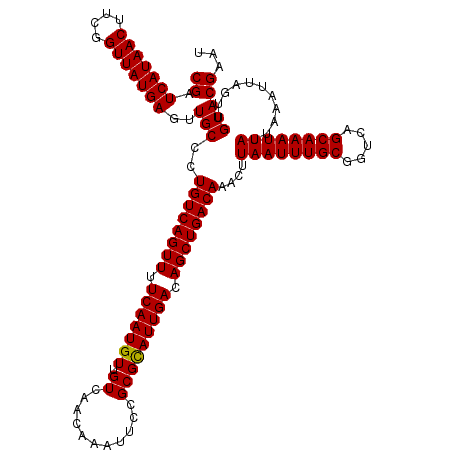
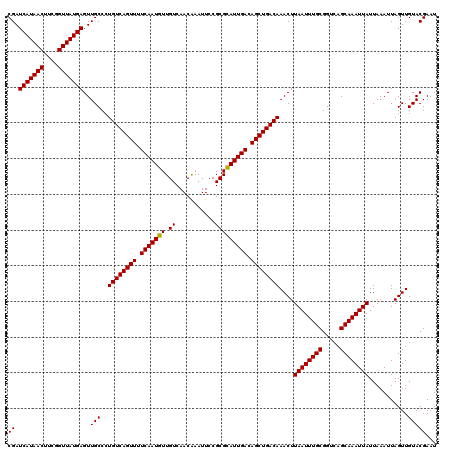
| Location | 382,705 – 382,820 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.90 |
| Mean single sequence MFE | -35.01 |
| Consensus MFE | -32.09 |
| Energy contribution | -32.40 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 382705 115 - 22407834 UCAGU-----UAACUCCAGUUCAUAACUCAUUUCUGGGCUCGAUCAUAACUUCGGUUAUGAGUUGCCAUGUCAGUUUUCAAUGUUGUCAACAAAUUCCGCGUAUUGACAGCUGACAAACU ..(((-----(.....(((..............)))(((....(((((((....)))))))...))).((((((((.(((((((((....)))........)))))).)))))))))))) ( -30.94) >DroSec_CAF1 50343 115 - 1 UCAGU-----UAACUCCAGUUCAUAACUCAUUCCGGGACUCGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCAAUGUUGUCAACAAAUUCCGCGCAUUGACAGCUGACAAACU ..(((-----(.......................(((.(....(((((((....)))))))...))))((((((((.(((((((.((...........))))))))).)))))))))))) ( -33.60) >DroSim_CAF1 51033 115 - 1 UCAGU-----UAACUCCAGUUCAUAACUCAUUCCGGGGCUCGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCAAUGUUGUCAACAAAUUCCGCGCAUUGACAGCUGACAAACU ..(((-----(.......................(((((....(((((((....)))))))...)))))(((((((.(((((((.((...........))))))))).))))))).)))) ( -37.70) >DroYak_CAF1 51647 120 - 1 GCAGCGUCGCAAACUCCAGUUCAUAACUCAUUCCUGGGCUCGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCAAUGUUGUCAACGAAUUCCGCGCAUUGACAGCUGACAAACU (((((.........((.((((((...........)))))).))(((((((....))))))))))))..((((((((.(((((((.((...........))))))))).)))))))).... ( -37.80) >consensus UCAGU_____UAACUCCAGUUCAUAACUCAUUCCGGGGCUCGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCAAUGUUGUCAACAAAUUCCGCGCAUUGACAGCUGACAAACU .................((((..............((((....(((((((....)))))))...))))((((((((.(((((((.((...........))))))))).)))))))))))) (-32.09 = -32.40 + 0.31)

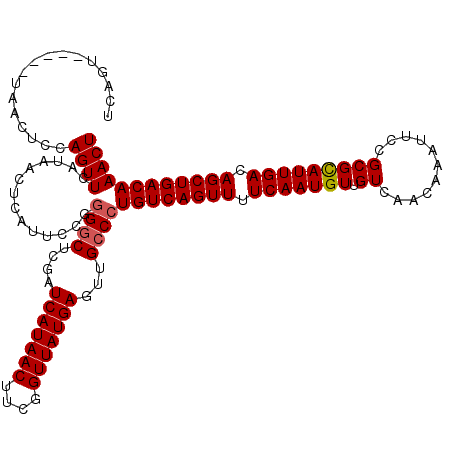
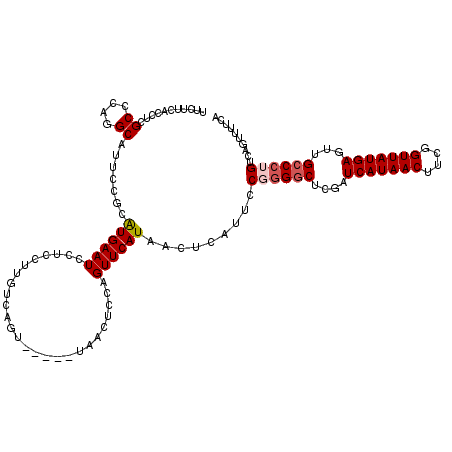
| Location | 382,745 – 382,860 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.20 |
| Mean single sequence MFE | -25.82 |
| Consensus MFE | -20.26 |
| Energy contribution | -21.07 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 382745 115 - 22407834 UUCCUCACCUCGCCCAAGCAUUCCGCAUGAAUCCUCCUUGUCAGU-----UAACUCCAGUUCAUAACUCAUUUCUGGGCUCGAUCAUAACUUCGGUUAUGAGUUGCCAUGUCAGUUUUCA ...........(((((.((.....))((((((.....(((.....-----))).....))))))..........)))))....(((((((....)))))))................... ( -22.30) >DroSec_CAF1 50383 115 - 1 UUCUUCACCUCGCCCAGGCAUUCCGCAUGAAUCCUCCUUGUCAGU-----UAACUCCAGUUCAUAACUCAUUCCGGGACUCGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCA ........((.((...((((.......(((.((((....((.(((-----((((....))...))))).))...)))).))).(((((((....)))))))..))))..)).))...... ( -23.20) >DroSim_CAF1 51073 115 - 1 UUCUUCACCUCGCCCAGGCAUUCCGCGUGAAUCCUCCUUGUCAGU-----UAACUCCAGUUCAUAACUCAUUCCGGGGCUCGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCA ...........((....))........((((..((.......(((-----((((....))...))))).....((((((....(((((((....)))))))...))))))..))..)))) ( -27.90) >DroYak_CAF1 51687 120 - 1 UUCUUCACCUCGCCCAGGCAUUUCGCAUGAAUCUUCCUCGGCAGCGUCGCAAACUCCAGUUCAUAACUCAUUCCUGGGCUCGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCA ...........(((((((........((((((........((......))........))))))........)))))))....(((((((....)))))))................... ( -29.88) >consensus UUCUUCACCUCGCCCAGGCAUUCCGCAUGAAUCCUCCUUGUCAGU_____UAACUCCAGUUCAUAACUCAUUCCGGGGCUCGAUCAUAACUUCGGUUAUGAGUUGCCCUGUCAGUUUUCA ...........((....)).......((((((..........................)))))).........((((((....(((((((....)))))))...)))))).......... (-20.26 = -21.07 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:26:40 2006