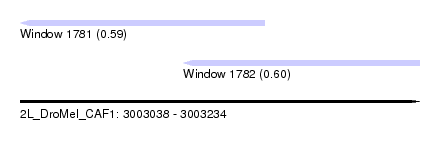
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 3,003,038 – 3,003,234 |
| Length | 196 |
| Max. P | 0.601298 |

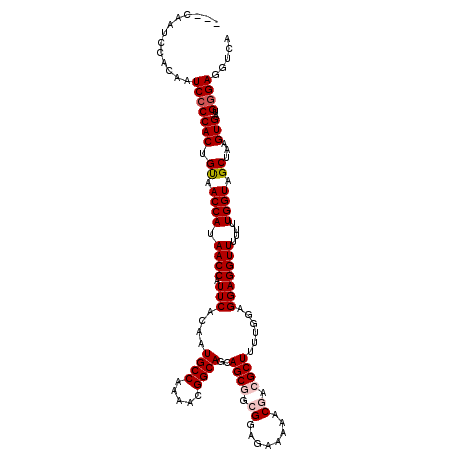
| Location | 3,003,038 – 3,003,158 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.17 |
| Mean single sequence MFE | -29.78 |
| Consensus MFE | -28.36 |
| Energy contribution | -28.60 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.593560 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3003038 120 - 22407834 UUGGAGGAGGUUUUUUUGGUAGCUAAGUGAUGGGAGGUCAAAUCAGUUACAGUUACGUGAGGCUGUUUUGGGCCAACAGGAGAACGUUUUGGCCAGAAGCAGAGCGAAUUGAAAGUAAAG (..(((((...)))))..)....(((.((((..(....)..)))).)))(((((.(((....(((((((.((((((..(.....)...)))))).))))))).))))))))......... ( -29.90) >DroSec_CAF1 10598 120 - 1 UUGGAGGAGGUUUUUUUGGUAGCUAAGUGAUGGGAGGUCAAAUCAGUUACAGUUACGUGAGGCUGUUUUGGGCCAACAGGAGAACGUUUUGACCAGAAGCAGAGCGAAUGGAAAGUAAAG ....((((.(((((((((((((((((.((((..(....)..)))).))..))))))....((((......))))..))))))))).))))..(((...((...))...)))......... ( -30.50) >DroSim_CAF1 9954 120 - 1 UUGGAGGAGGUUUUUUUGGUAGCUAAGUGAUGGGAGGUCAAAUCAGUUACAGUUACGUGAGGCUGUUUUGGGCCAACAGGAGAACGUUUUGGCCAGAAGCAGAGCGAAUGGAAAGUAAAG ....((((.(((((((((((((((((.((((..(....)..)))).))..))))))....((((......))))..))))))))).))))..(((...((...))...)))......... ( -30.20) >DroEre_CAF1 10621 119 - 1 UUGGAGGAGGUUU-UUUGGUAGCUAAGUGAUGGGAGGUCAAAUCAGUUACAGCUACGUGAGGCUGUUUUGGGCCAACAGGAGAACGUUUUGGGCAGAAGCAGAGCGAAUGGAAAGUAAAG ..........(((-((((((((((((.((((..(....)..)))).))..))))))....((((......))))..))))))).(((((((........))))))).............. ( -29.00) >DroYak_CAF1 10189 119 - 1 UUGGAGGAGGUUU-UUUGGUAGCUAAGUGAUGGGAGGUCAAAUCAGUUACAGUUACGUGAGGCUGUUUUGGGCCAACAGGAGAACGUUUUGGCCAGAAGCAGAGCGAAUGGAAAGUAAAG ..........(((-(((.((((((...((((.....))))....)))))).....(((....(((((((.((((((..(.....)...)))))).))))))).)))...))))))..... ( -29.30) >consensus UUGGAGGAGGUUUUUUUGGUAGCUAAGUGAUGGGAGGUCAAAUCAGUUACAGUUACGUGAGGCUGUUUUGGGCCAACAGGAGAACGUUUUGGCCAGAAGCAGAGCGAAUGGAAAGUAAAG .........(((((((((((((((((.((((..(....)..)))).))..))))))....((((......))))..)))))))))((((((........))))))............... (-28.36 = -28.60 + 0.24)

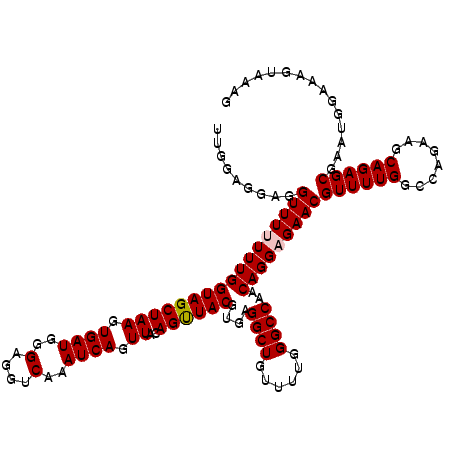
| Location | 3,003,118 – 3,003,234 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 93.66 |
| Mean single sequence MFE | -34.04 |
| Consensus MFE | -26.32 |
| Energy contribution | -27.16 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 3003118 116 - 22407834 ---CAAUCCGCAAUCCCCACUGUAACCAUAACCAUUCACAAUGCCAAAACGGCAGCAGCGGCGGAAAAAAACGACGCUUUUGGAGGAGGUUUUUUUGGUAGCUAAGUGAUGGGAGGUCA ---..........((((((((((.((((.((((.(((.(((((((.....))))..((((.((........)).)))).))))))..))))....)))).))..))))..))))..... ( -32.70) >DroSec_CAF1 10678 116 - 1 ---CAAUCCACAAUCCCCACAGUAACCAUAACCAUUCAAAAUGCCAAAACGGCAGCAGCGGCGGAGAAAAACGACGCUUUUGGAGGAGGUUUUUUUGGUAGCUAAGUGAUGGGAGGUCA ---..........((((((((((.((((.((((.(((((((((((.....))))...(((.((........)).))))))))))...))))....)))).)))..)))..))))..... ( -33.60) >DroSim_CAF1 10034 116 - 1 ---CAAUCCACAAUCCCCACAGCAACCAUAACCAUUCACAAUGCCAAAACGGCAGCAGCGGCGGAGAAAAACGACGCUUUUGGAGGAGGUUUUUUUGGUAGCUAAGUGAUGGGAGGUCA ---..........((((((((((.((((.((((.(((....((((.....)))).(((.((((..(.....)..)))).)))..)))))))....)))).)))..)))..))))..... ( -34.20) >DroEre_CAF1 10701 112 - 1 ---CGAUCCCCAAUCACCACUGUAACCAUAACCAUUCACAAUGCCAAAACGGCAGCAGC---GGAGAAAAACGGCGCUUUUGGAGGAGGUUU-UUUGGUAGCUAAGUGAUGGGAGGUCA ---.((((((((.((((....((.((((.((((.(((.(((((((.....))))((.((---.(.......).))))..))))))..)))).-..)))).))...)))))))).)))). ( -37.70) >DroYak_CAF1 10269 115 - 1 ACCCAAUCCCCAAUCCCCACUGUAACCAUAACCAUUCACAAUGCCAAAACGGCAGCAGC---GGAGAAAAACGACGCUUUUGGAGGAGGUUU-UUUGGUAGCUAAGUGAUGGGAGGUCA .........((..((((((((((.((((.((((.(((.(((((((.....))))..(((---(..(.....)..)))).))))))..)))).-..)))).))..))))..))))))... ( -32.00) >consensus ___CAAUCCACAAUCCCCACUGUAACCAUAACCAUUCACAAUGCCAAAACGGCAGCAGCGGCGGAGAAAAACGACGCUUUUGGAGGAGGUUUUUUUGGUAGCUAAGUGAUGGGAGGUCA .............(((((((.((.((((.((((.(((....((((.....))))..((((.((........)).))))......)))))))....)))).))...)))..))))..... (-26.32 = -27.16 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:53:32 2006