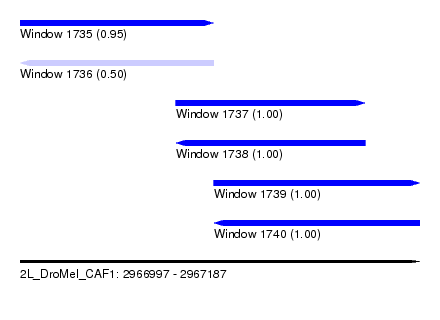
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 2,966,997 – 2,967,187 |
| Length | 190 |
| Max. P | 0.999532 |

| Location | 2,966,997 – 2,967,089 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -38.58 |
| Consensus MFE | -25.98 |
| Energy contribution | -25.03 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.22 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.945374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2966997 92 + 22407834 GUUUCUCGAUGCACCUUCGCGUUCUGAAGCCAAUAUGGUUGUGCAAAUGCCUGCAGACGCC--ACCGGAAACGGAAAC----------GUCUGCAGAGCGCCCU (((((..(((((......)))))..)))))......((.(((((....))(((((((((..--.(((....)))...)----------)))))))).))).)). ( -36.90) >DroPse_CAF1 127400 104 + 1 GUUUCUAGAGGCACAUUCGCGUUUUGGAGCCAAUAAGGUUGUGCAAUUGCCUGCAGGUUUCUAAUCGGAAGCGGAAGCGAACGUGGAAGCCUGCAGCGCCCUCU ......(((((((((..(.....(((....)))...)..)))))....(((((((((((((((..((...((....))...))))))))))))))).)).)))) ( -43.40) >DroSim_CAF1 20092 92 + 1 GUUUCUCGAUGCACCUUCGCGUUCUGAAGCAAAUAUGGUUGUGCAAAUGCCUGCAGACGCU--ACCGGAAACGGAAAC----------GUCUGCAGAGCGCCCU (((((..(((((......)))))..)))))......((.(((((....))(((((((((..--.(((....)))...)----------)))))))).))).)). ( -37.80) >DroEre_CAF1 19487 88 + 1 --UUCUCGAGGCACCUUCGCGUUCUGAAGCCAAUAAGGUUGUGCAAAUGCCUGCAGACGCC--ACCGGAAACGGAA------------GUCUGCAGAGCGCCCU --.......(((......(((((.((.((((.....))))...)))))))((((((((...--.(((....)))..------------))))))))...))).. ( -35.00) >DroYak_CAF1 19628 88 + 1 --UUCUCGAGGCACCUUCGCGUUCUGAAGCCAAUAAGGUUGUGCAAAUGCCUGCAGACGCC--ACCGGAAACGGAA------------GUCUGCAGAGCGCCCU --.......(((......(((((.((.((((.....))))...)))))))((((((((...--.(((....)))..------------))))))))...))).. ( -35.00) >DroPer_CAF1 129005 104 + 1 GUUUCUAGAGGCACAUUCGCGUUUUGGAGCCAAUAAGGUUGUGCAAUUGCCUGCAGGUUUCUAAUCGGAAGCGGAAGCGAACGUGGAAGCCUGCAGCGCCCUCU ......(((((((((..(.....(((....)))...)..)))))....(((((((((((((((..((...((....))...))))))))))))))).)).)))) ( -43.40) >consensus GUUUCUCGAGGCACCUUCGCGUUCUGAAGCCAAUAAGGUUGUGCAAAUGCCUGCAGACGCC__ACCGGAAACGGAA_C__________GUCUGCAGAGCGCCCU ..........((((..(((.....)))((((.....))))))))....((((((((((......(((....)))..............)))))))).))..... (-25.98 = -25.03 + -0.94)

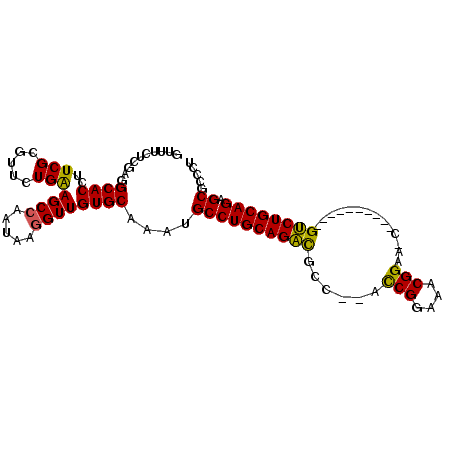
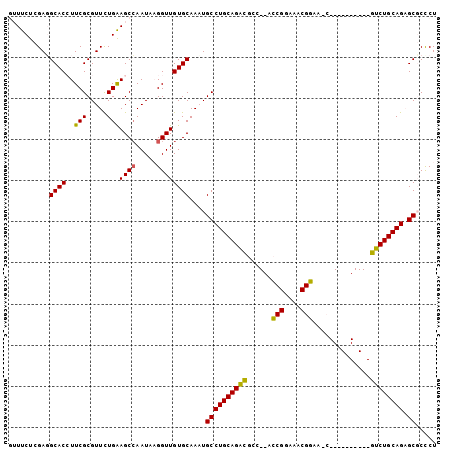
| Location | 2,966,997 – 2,967,089 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 81.27 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -20.13 |
| Energy contribution | -20.47 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2966997 92 - 22407834 AGGGCGCUCUGCAGAC----------GUUUCCGUUUCCGGU--GGCGUCUGCAGGCAUUUGCACAACCAUAUUGGCUUCAGAACGCGAAGGUGCAUCGAGAAAC .....((.((((((((----------((..(((....))).--.))))))))))))...(((((..((.....))((((.......)))))))))......... ( -37.30) >DroPse_CAF1 127400 104 - 1 AGAGGGCGCUGCAGGCUUCCACGUUCGCUUCCGCUUCCGAUUAGAAACCUGCAGGCAAUUGCACAACCUUAUUGGCUCCAAAACGCGAAUGUGCCUCUAGAAAC (((((((.(((((((.(((..((...((....))...))....))).))))))))).............((((.((........)).))))..)))))...... ( -29.00) >DroSim_CAF1 20092 92 - 1 AGGGCGCUCUGCAGAC----------GUUUCCGUUUCCGGU--AGCGUCUGCAGGCAUUUGCACAACCAUAUUUGCUUCAGAACGCGAAGGUGCAUCGAGAAAC .....((.((((((((----------(((.(((....))).--)))))))))))))...(((((.......(((((........))))).)))))......... ( -35.90) >DroEre_CAF1 19487 88 - 1 AGGGCGCUCUGCAGAC------------UUCCGUUUCCGGU--GGCGUCUGCAGGCAUUUGCACAACCUUAUUGGCUUCAGAACGCGAAGGUGCCUCGAGAA-- .((((((.((((((((------------..(((....))).--...)))))))))).........(((((....((........)).)))))))))......-- ( -31.20) >DroYak_CAF1 19628 88 - 1 AGGGCGCUCUGCAGAC------------UUCCGUUUCCGGU--GGCGUCUGCAGGCAUUUGCACAACCUUAUUGGCUUCAGAACGCGAAGGUGCCUCGAGAA-- .((((((.((((((((------------..(((....))).--...)))))))))).........(((((....((........)).)))))))))......-- ( -31.20) >DroPer_CAF1 129005 104 - 1 AGAGGGCGCUGCAGGCUUCCACGUUCGCUUCCGCUUCCGAUUAGAAACCUGCAGGCAAUUGCACAACCUUAUUGGCUCCAAAACGCGAAUGUGCCUCUAGAAAC (((((((.(((((((.(((..((...((....))...))....))).))))))))).............((((.((........)).))))..)))))...... ( -29.00) >consensus AGGGCGCUCUGCAGAC__________G_UUCCGUUUCCGGU__GGCGUCUGCAGGCAUUUGCACAACCUUAUUGGCUUCAGAACGCGAAGGUGCCUCGAGAAAC .(((.((.((((((((..............(((....)))......))))))))))....((((.......((.((........)).)).)))))))....... (-20.13 = -20.47 + 0.34)
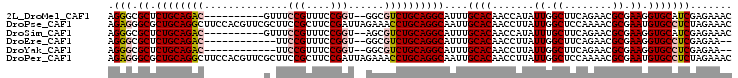


| Location | 2,967,071 – 2,967,161 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -39.62 |
| Consensus MFE | -31.42 |
| Energy contribution | -33.30 |
| Covariance contribution | 1.88 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.15 |
| Structure conservation index | 0.79 |
| SVM decision value | 3.29 |
| SVM RNA-class probability | 0.998937 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2967071 90 + 22407834 ACGUCUGCAGAGCGCCCUCUUUAGGCGGCUUGUAAACG--------AUGCAAACGUGCAACCCUGCAACCGCUGCAUCGUUUACACUCGGCUUGAAAU .((((((.((((....)))).))))))(((((((((((--------(((((....((((....)))).....)))))))))))))...)))....... ( -36.20) >DroSec_CAF1 28223 98 + 1 ACGUCCGCAGUGCGCCCUCUUUAGGCGGCUUGUAAGCGAUGCAGCUAUGCAAACAUGCAACCCUGCAACCGCUGCAUCGUUUACACUCGGCUUGAAAU ..(..(((...)))..)..(((((((.(..(((((((((((((((..((((............))))...)))))))))))))))..).))))))).. ( -38.10) >DroSim_CAF1 20166 98 + 1 ACGUCUGCAGAGCGCCCUCUUUAGGCGGCUUGUAAGCGGUGCAGCGAUGCAAACGUGCAACCCUGCAACCGCUGCAUCGUUUACACUCGGCUUGAAAU .((((((.((((....)))).))))))(((((((((((((((((((.((((............))))..))))))))))))))))...)))....... ( -42.60) >DroEre_CAF1 19559 96 + 1 --GUCUGCAGAGCGCCCUCUUUAGGCGGCUUGUAAACGAUGCAGCGAUGCAACCGUGCCACCCUGCAACCGCUGCAUCGUUUACACUCAGCUUGAAAU --(((((.((((....)))).)))))((((((((((((((((((((.((((............))))..))))))))))))))))...))))...... ( -41.50) >DroYak_CAF1 19700 96 + 1 --GUCUGCAGAGCGCCCUCUUCAGGCGGCUUGUAAACGAUGCAGCGAUCCAAUCGGGCAACCCUGCAACCGCUGCACCGUUUACACUCAGCUUGAAAU --(((((.((((....)))).)))))((((((((((((.(((((((..((....))(((....)))...))))))).))))))))...))))...... ( -39.70) >consensus ACGUCUGCAGAGCGCCCUCUUUAGGCGGCUUGUAAACGAUGCAGCGAUGCAAACGUGCAACCCUGCAACCGCUGCAUCGUUUACACUCGGCUUGAAAU ..(((((.((((....)))).)))))((((((((((((((((((((.((((............))))..))))))))))))))))...))))...... (-31.42 = -33.30 + 1.88)

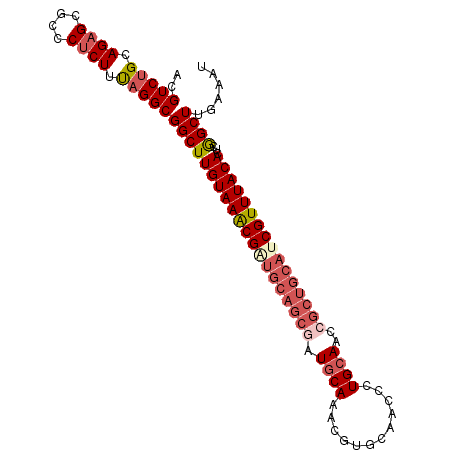
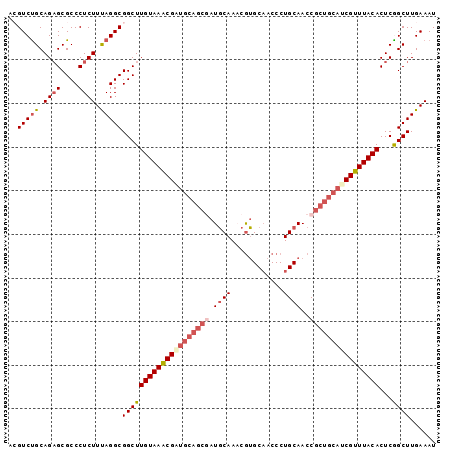
| Location | 2,967,071 – 2,967,161 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 89.67 |
| Mean single sequence MFE | -41.06 |
| Consensus MFE | -34.28 |
| Energy contribution | -37.40 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.39 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2967071 90 - 22407834 AUUUCAAGCCGAGUGUAAACGAUGCAGCGGUUGCAGGGUUGCACGUUUGCAU--------CGUUUACAAGCCGCCUAAAGAGGGCGCUCUGCAGACGU .......((.(((((((((((((((((((..(((......))))).))))))--------))))))).....((((.....)))))))).))...... ( -36.50) >DroSec_CAF1 28223 98 - 1 AUUUCAAGCCGAGUGUAAACGAUGCAGCGGUUGCAGGGUUGCAUGUUUGCAUAGCUGCAUCGCUUACAAGCCGCCUAAAGAGGGCGCACUGCGGACGU .......(((((((((((.(((((((((...((((((........))))))..))))))))).))))..((.((((.....))))))))).))).).. ( -39.80) >DroSim_CAF1 20166 98 - 1 AUUUCAAGCCGAGUGUAAACGAUGCAGCGGUUGCAGGGUUGCACGUUUGCAUCGCUGCACCGCUUACAAGCCGCCUAAAGAGGGCGCUCUGCAGACGU .......((.((((((((.((.(((((((((.(((((........)))))))))))))).)).)))).....((((.....)))))))).))...... ( -37.80) >DroEre_CAF1 19559 96 - 1 AUUUCAAGCUGAGUGUAAACGAUGCAGCGGUUGCAGGGUGGCACGGUUGCAUCGCUGCAUCGUUUACAAGCCGCCUAAAGAGGGCGCUCUGCAGAC-- .......((.(((((((((((((((((((((.((((..........))))))))))))))))))))).....((((.....)))))))).))....-- ( -45.20) >DroYak_CAF1 19700 96 - 1 AUUUCAAGCUGAGUGUAAACGGUGCAGCGGUUGCAGGGUUGCCCGAUUGGAUCGCUGCAUCGUUUACAAGCCGCCUGAAGAGGGCGCUCUGCAGAC-- .......((.((((((((((((((((((((((...(((...))).....)))))))))))))))))).....((((.....)))))))).))....-- ( -46.00) >consensus AUUUCAAGCCGAGUGUAAACGAUGCAGCGGUUGCAGGGUUGCACGUUUGCAUCGCUGCAUCGUUUACAAGCCGCCUAAAGAGGGCGCUCUGCAGACGU .......((.(((((((((((((((((((((.(((((........)))))))))))))))))))))).....((((.....)))))))).))...... (-34.28 = -37.40 + 3.12)


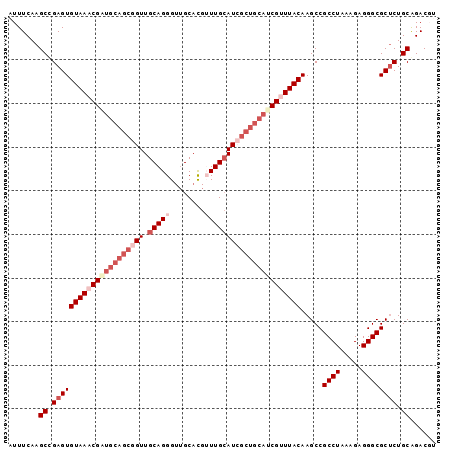
| Location | 2,967,089 – 2,967,187 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -39.66 |
| Consensus MFE | -33.40 |
| Energy contribution | -35.12 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -5.07 |
| Structure conservation index | 0.84 |
| SVM decision value | 3.48 |
| SVM RNA-class probability | 0.999283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2967089 98 + 22407834 CUUUAGGCGGCUUGUAAACG--------AUGCAAACGUGCAACCCUGCAACCGCUGCAUCGUUUACACUCGGCUUGAAAUGUCUGUUUCUCUCACACAGAUCACAC .(((((((.(..((((((((--------(((((....((((....)))).....)))))))))))))..).)))))))..((((((.........))))))..... ( -35.60) >DroSec_CAF1 28241 106 + 1 CUUUAGGCGGCUUGUAAGCGAUGCAGCUAUGCAAACAUGCAACCCUGCAACCGCUGCAUCGUUUACACUCGGCUUGAAAUGUCUGUUUCUCUCACACAGAUCACAC .(((((((.(..(((((((((((((((..((((............))))...)))))))))))))))..).)))))))..((((((.........))))))..... ( -40.20) >DroSim_CAF1 20184 106 + 1 CUUUAGGCGGCUUGUAAGCGGUGCAGCGAUGCAAACGUGCAACCCUGCAACCGCUGCAUCGUUUACACUCGGCUUGAAAUGUCUGUUUCUCUCACACAGAUCACAC .(((((((.(..((((((((((((((((.((((............))))..))))))))))))))))..).)))))))..((((((.........))))))..... ( -42.00) >DroEre_CAF1 19575 106 + 1 CUUUAGGCGGCUUGUAAACGAUGCAGCGAUGCAACCGUGCCACCCUGCAACCGCUGCAUCGUUUACACUCAGCUUGAAAUGUCUGUUUCUCUCACACAGAUCACAC .(((((((....((((((((((((((((.((((............))))..))))))))))))))))....)))))))..((((((.........))))))..... ( -40.90) >DroYak_CAF1 19716 106 + 1 CUUCAGGCGGCUUGUAAACGAUGCAGCGAUCCAAUCGGGCAACCCUGCAACCGCUGCACCGUUUACACUCAGCUUGAAAUGUCUGUUUCUCUCACACAGAUCACAC .(((((((....((((((((.(((((((..((....))(((....)))...))))))).))))))))....)))))))..((((((.........))))))..... ( -39.60) >consensus CUUUAGGCGGCUUGUAAACGAUGCAGCGAUGCAAACGUGCAACCCUGCAACCGCUGCAUCGUUUACACUCGGCUUGAAAUGUCUGUUUCUCUCACACAGAUCACAC .(((((((.(..((((((((((((((((.((((............))))..))))))))))))))))..).)))))))..((((((.........))))))..... (-33.40 = -35.12 + 1.72)

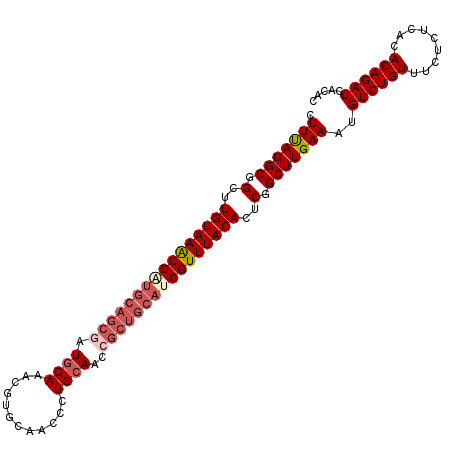

| Location | 2,967,089 – 2,967,187 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -38.72 |
| Consensus MFE | -32.32 |
| Energy contribution | -35.24 |
| Covariance contribution | 2.92 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.79 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 2967089 98 - 22407834 GUGUGAUCUGUGUGAGAGAAACAGACAUUUCAAGCCGAGUGUAAACGAUGCAGCGGUUGCAGGGUUGCACGUUUGCAU--------CGUUUACAAGCCGCCUAAAG ...(((..(((.((.......)).)))..))).((.(..((((((((((((((((..(((......))))).))))))--------))))))))..).))...... ( -33.50) >DroSec_CAF1 28241 106 - 1 GUGUGAUCUGUGUGAGAGAAACAGACAUUUCAAGCCGAGUGUAAACGAUGCAGCGGUUGCAGGGUUGCAUGUUUGCAUAGCUGCAUCGCUUACAAGCCGCCUAAAG ...(((..(((.((.......)).)))..))).((.(..(((((.(((((((((...((((((........))))))..))))))))).)))))..).))...... ( -36.70) >DroSim_CAF1 20184 106 - 1 GUGUGAUCUGUGUGAGAGAAACAGACAUUUCAAGCCGAGUGUAAACGAUGCAGCGGUUGCAGGGUUGCACGUUUGCAUCGCUGCACCGCUUACAAGCCGCCUAAAG ...(((..(((.((.......)).)))..))).((.(..(((((.((.(((((((((.(((((........)))))))))))))).)).)))))..).))...... ( -34.80) >DroEre_CAF1 19575 106 - 1 GUGUGAUCUGUGUGAGAGAAACAGACAUUUCAAGCUGAGUGUAAACGAUGCAGCGGUUGCAGGGUGGCACGGUUGCAUCGCUGCAUCGUUUACAAGCCGCCUAAAG ...(((..(((.((.......)).)))..))).((.(..((((((((((((((((((.((((..........))))))))))))))))))))))..).))...... ( -42.20) >DroYak_CAF1 19716 106 - 1 GUGUGAUCUGUGUGAGAGAAACAGACAUUUCAAGCUGAGUGUAAACGGUGCAGCGGUUGCAGGGUUGCCCGAUUGGAUCGCUGCAUCGUUUACAAGCCGCCUGAAG ..(((.(((((.........))))))))((((.((.(..(((((((((((((((((((...(((...))).....)))))))))))))))))))..).)).)))). ( -46.40) >consensus GUGUGAUCUGUGUGAGAGAAACAGACAUUUCAAGCCGAGUGUAAACGAUGCAGCGGUUGCAGGGUUGCACGUUUGCAUCGCUGCAUCGUUUACAAGCCGCCUAAAG ...(((..(((.((.......)).)))..))).((.(..((((((((((((((((((.(((((........)))))))))))))))))))))))..).))...... (-32.32 = -35.24 + 2.92)

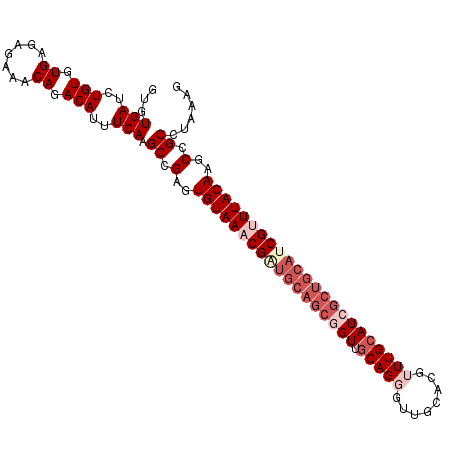
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:52:52 2006