
| Window 2 | |
|
| Window 1 | |
|

| Location | 0 – 120 |
|---|---|
| Length | 120 |
| RNAz 1.0 Sequences | 2 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 74.17 |
| Mean single sequence MFE | -58.45 |
| Consensus MFE | -38.15 |
| Energy contribution | -39.40 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
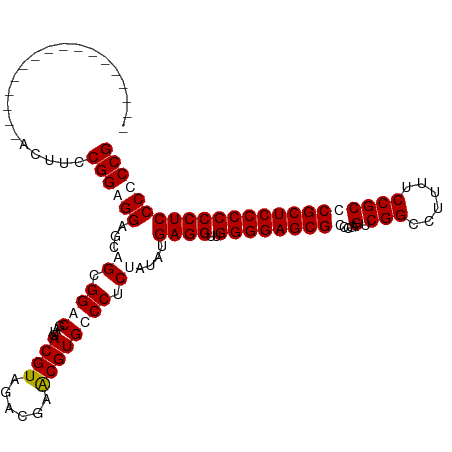
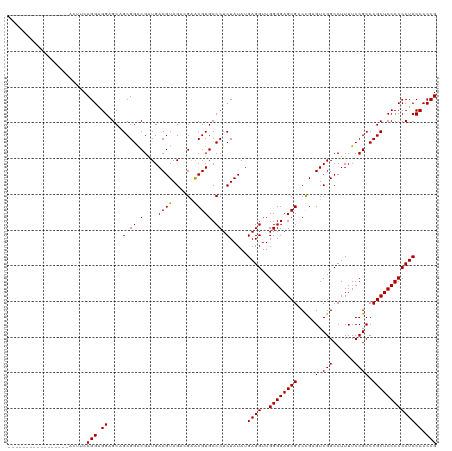
>Mus_1_466/0-120 -----------------AGUUCCGGGGGAGCCGCGGACGGUGACGUAGCCGAGCGUGCCCUCUAUAUGAGGUUGGGGAGCGCCCGCGUCGGCCUUUUCCGCCCGCUCCCCCCUCCCCCCG -----------------......((((((((.(((((((....))).(((((.(((..((((.....))))..(((.....))))))))))).....))))..))))))))......... ( -51.30) >Homo_1_499/0-120 ACGUCACCGCCUGCGUCGCUUCCGGAGGCGCAGCGGGCGAUGACGUAGAGGGACGUGCCCUCUAUAUGAGGUUGGGGAGCGGCUGAGUCGGCCUUUUCCGCCCGCUCCCCCCUCCCCCCG ((((((.(((((((..(((((....)))))..))))))).))))))((((((.....))))))....((((..((((((((((.(((........))).).)))))))))))))...... ( -65.60) >consensus _________________ACUUCCGGAGGAGCAGCGGACGAUGACGUAGACGAACGUGCCCUCUAUAUGAGGUUGGGGAGCGCCCGAGUCGGCCUUUUCCGCCCGCUCCCCCCUCCCCCCG ......................((((((......((((....((((......)))))))).......((((..(((((((((....(.(((......))))))))))))))))))))))) (-38.15 = -39.40 + 1.25)

| Location | 0 – 120 |
|---|---|
| Length | 120 |
| Sequences | 2 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.17 |
| Mean single sequence MFE | -61.15 |
| Consensus MFE | -45.30 |
| Energy contribution | -45.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
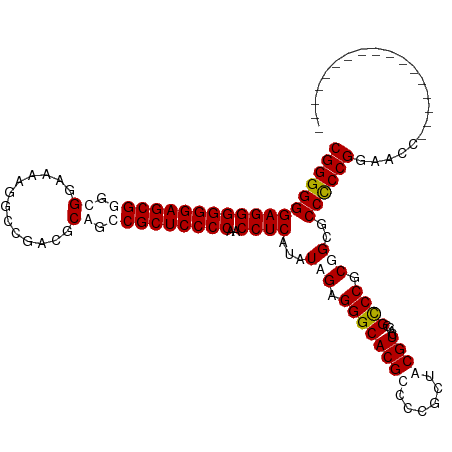
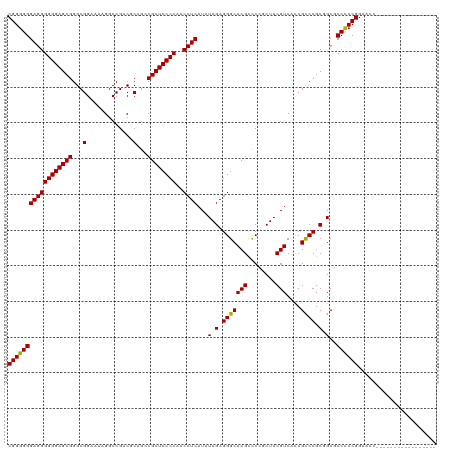
>Mus_1_466/0-120 CGGGGGGAGGGGGGAGCGGGCGGAAAAGGCCGACGCGGGCGCUCCCCAACCUCAUAUAGAGGGCACGCUCGGCUACGUCACCGUCCGCGGCUCCCCCGGAACU----------------- (.(((((((......((((((((....((((((.(((((.....)))..((((.....))))....))))))))......))))))))..))))))).)....----------------- ( -53.50) >Homo_1_499/0-120 CGGGGGGAGGGGGGAGCGGGCGGAAAAGGCCGACUCAGCCGCUCCCCAACCUCAUAUAGAGGGCACGUCCCUCUACGUCAUCGCCCGCUGCGCCUCCGGAAGCGACGCAGGCGGUGACGU .(((((((.((((((((((.(((......)))......)))))))))..((((.....))))...).)))))))(((((((((((.((..(((........)))..)).))))))))))) ( -68.80) >consensus CGGGGGGAGGGGGGAGCGGGCGGAAAAGGCCGACGCAGCCGCUCCCCAACCUCAUAUAGAGGGCACGCCCCGCUACGUCACCGCCCGCGGCGCCCCCGGAACC_________________ (((((((((((((((((((.(((......)))......)))))))))..)))).......(((((((........)))....))))......))))))...................... (-45.30 = -45.30 + 0.00)

These windows have a probability below the cutoff.
| Window | Location | Strand | N | Length | Mean pairwise ID | z | SCI | P |
| 3 | 40 – 160 | + | 2 | 120 | 85.00 | -0.90 | 0.80 | 0.126295 |
| 4 | 40 – 160 | - | 2 | 120 | 85.00 | -1.13 | 0.84 | 0.244517 |
| 5 | 120 – 240 | + | 6 | 120 | 80.24 | 1.07 | 0.65 | 0.000003 |
| 6 | 120 – 240 | - | 6 | 120 | 80.24 | 0.51 | 0.69 | 0.006145 |
| 7 | 160 – 280 | + | 6 | 120 | 85.29 | 0.56 | 0.64 | 0.059107 |
| 8 | 160 – 280 | - | 6 | 120 | 85.29 | 0.51 | 0.64 | 0.060730 |
| 9 | 200 – 320 | + | 6 | 120 | 73.17 | -0.80 | 0.38 | 0.000001 |
| 10 | 200 – 320 | - | 6 | 120 | 73.17 | -0.30 | 0.30 | 0.000000 |
| 11 | 240 – 360 | + | 6 | 120 | 81.61 | -1.72 | 0.51 | 0.184281 |
| 12 | 240 – 360 | - | 6 | 120 | 81.61 | -1.33 | 0.32 | 0.000004 |
| 13 | 280 – 400 | + | 6 | 120 | 84.83 | -2.13 | 0.65 | 0.356793 |
| 14 | 280 – 400 | - | 6 | 120 | 84.83 | -0.68 | 0.43 | 0.005290 |
| 15 | 320 – 440 | + | 6 | 120 | 84.50 | -1.05 | 0.56 | 0.093609 |
| 16 | 320 – 440 | - | 6 | 120 | 84.50 | -1.32 | 0.51 | 0.050108 |
| 17 | 360 – 480 | + | 6 | 120 | 77.78 | -0.43 | 0.27 | 0.000000 |
| 18 | 360 – 480 | - | 6 | 120 | 77.78 | -0.80 | 0.40 | 0.000001 |
| 19 | 382 – 502 | + | 6 | 120 | 85.44 | -0.51 | 0.53 | 0.053838 |
| 20 | 382 – 502 | - | 6 | 120 | 85.44 | 0.47 | 0.59 | 0.069272 |