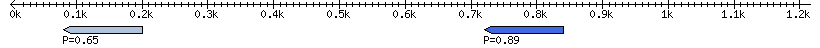
| Window 34 | |
|
| Window 6 | |
|
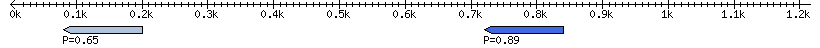
| Location | 80 – 200 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.48 |
| Mean single sequence MFE | -52.08 |
| Consensus MFE | -35.85 |
| Energy contribution | -39.05 |
| Covariance contribution | 3.20 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
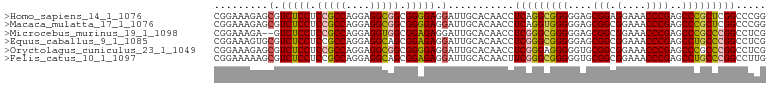
>Equus_caballus_9_1_1085/80-200 GGGGGAGCGGGCGGAAAA-GCCGACUCAGCCGCUCCCCAACCUCAUAUAGA-GGCACGUCCACCUACGUCAUCGCCCGCUGCGCCUCCGGAAGUUCCGCAAGCGGUGACGUGG .(((((((((.(((....-.)))......)))))))))..((((.....))-)).........((((((((((((....((((....(....)...)))).)))))))))))) ( -51.80) >Oryctolagus_cuniculus_23_1_1049/80-200 GGGGGAGCGGGCGGAAAAGGCCGACUCAGCCGCUCCCCAACCUCAUAUAGAGGGCACGUCCUUCUACGUCAUCGCCCGCUGCGCUCCCGGAAGCGCCGCAAGCAGUGACGUGG .(((((((((.(((......)))......)))))))))..((((.....))))..........(((((((((.((..((.(((((......))))).))..)).))))))))) ( -54.50) >Mus_musculus_21_1_983/80-200 GGGGGAGCGGGCGGAAAAGGCCGACGCGGGCGCUCCCCAACCUCAUAUAGAGGGCACGCUCGGCUACGUCACCGUCCGCGGCUCCCCCGGAACUGUCGUCAGCGGUGACGUGA ((((((((((((((....((((((.(((((.....)))..((((.....))))....))))))))......))))))...))))))))(..(((((.....)))))..).... ( -56.00) >Rattus_norvegicus_30_1_942/80-200 GGGGGAGCGGGCGGAAAAGGCCGACUCGGGCGCUUCCCAACCUCAUAUAGAGGGCACGCUCAUCUACGUCAUCGCCCGCUGCGCCCCCGGAACUGUCGUAAGCGGUGACGUGG ((((((((((((((((..(.(((...))).)..))))...((((.....))))....................)))))))...)))))(..((((.(....)))))..).... ( -47.70) >Cavia_porcellus_6_1_1053/80-200 GGGGGAGCGGGCGGAAAAGGCCGACUCAGCCGCUCCCCAACCUCAUAUAGAGGGCACACUC--UUACGUCACCGCGUGCUGCGCCUCCGGAAGCGCCGUAAGCGUUGGCGUGG .(((((((((.(((......)))......)))))))))..((((.....))))........--.(((((((.(((.(((.((((........)))).))).))).))))))). ( -52.20) >Mus_1_466/80-200 GGGGGAGCGGGCGGAAAAGGCCGACGCGGGCGCUCCCCAACCUCAUAUAGAGGGCACGCUCGGCUACGUCACCGUCCGCGGCUCCCCCGGAACU------------------- ((((((((((((((....((((((.(((((.....)))..((((.....))))....))))))))......))))))...))))))))......------------------- ( -50.30) >consensus GGGGGAGCGGGCGGAAAAGGCCGACUCAGCCGCUCCCCAACCUCAUAUAGAGGGCACGCUCAGCUACGUCACCGCCCGCUGCGCCCCCGGAACUGCCGUAAGCGGUGACGUGG .(((((((((((((......))).))....))))))))..((((.....))))..........((((((((((((..((.(((..........))).))..)))))))))))) (-35.85 = -39.05 + 3.20)

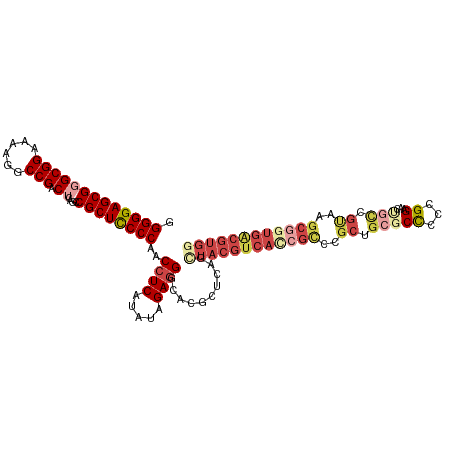
| Location | 720 – 840 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 91.88 |
| Mean single sequence MFE | -46.03 |
| Consensus MFE | -40.49 |
| Energy contribution | -40.55 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891835 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
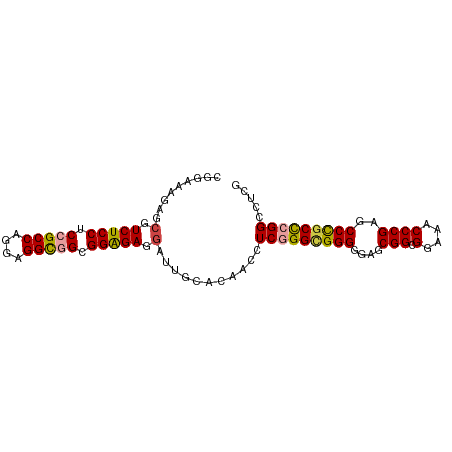
>Homo_sapiens_14_1_1076/720-840 CGGAAAGAGCGUCUCCUCCGCCAGGAGGCGGCGGGGAGGAUUGCACAACCUCAGGCGGGGGAGCGGAGGAAACCCGAGCCCGCUCGGCCCGG (((...(((((.((((((((((..((((..((((......))))....)))).))))))))))(((.(....))))....)))))...))). ( -48.40) >Macaca_mulatta_17_1_1076/720-840 CGGAAAGAGCGUCUCCUCCGCCAGGAGGCGGCGGGGAGGAUUGCACAACCUCAGGUGGGGGAGCGGCGGAAACCCGAGCCCGCUCGGCCCGG (((...(((((.((((((((((..((((..((((......))))....)))).))))))))))(((.(....))))....)))))...))). ( -46.50) >Microcebus_murinus_19_1_1098/720-840 CGGAAAGA--GUCUCCUCCGCCAGGAGGUGGCGGAGAGGAUUGCACAACCUCGGGCGGGGGAGCGGCGGAAACCCGAGCCCGCCCGGCCUCG .((....(--(((..((((((((.....))))))))..))))........(((((((((....(((.(....))))..)))))))))))... ( -47.80) >Equus_caballus_9_1_1085/720-840 CGGAAAGUGCGUCUCCUCCGCCAGGAGGCAGCGGAGAGGAUUGCACAACCUCGGGCGGGGGAGCGGCGGAAACCCGAGCCUGCCCGGCCUCG .((...(((((((..((((((.........))))))..)).)))))....(((((((((....(((.(....))))..)))))))))))... ( -46.20) >Oryctolagus_cuniculus_23_1_1049/720-840 CGGAAAGAGCGUCUCCUCCGCCAGGAGGCGGCGGGGAGGAUUGCACAACCUCGGGAGGGGGUGCGGCGGAAACCCGAGCCCGCCCGGCCUCG ......(((.(((.(((((....))))).((((((.......((((..(((....)))..))))((.(....)))...)))))).)))))). ( -45.30) >Felis_catus_10_1_1097/720-840 CGGAAAAAGCGUCUCCUCCGCCAGGAGGCAGCGGAGAGGAUUGCACAACUUCGGGCGGGGGUGCGGCGGAAACCCGAGCCUGCCCGGCCUUG .((.....(((((..((((((.........))))))..)).)))......(((((((((....(((.(....))))..)))))))))))... ( -42.00) >consensus CGGAAAGAGCGUCUCCUCCGCCAGGAGGCGGCGGAGAGGAUUGCACAACCUCGGGCGGGGGAGCGGCGGAAACCCGAGCCCGCCCGGCCUCG .((......(.(((((.(((((....))))).))))).)...........(((((((((....(((.(....))))..)))))))))))... (-40.49 = -40.55 + 0.06)
These windows have a probability below the cutoff.
| Window | Location | Strand | N | Length | Mean pairwise ID | z | SCI | P |
| 1 | 0 – 120 | + | 6 | 120 | 72.08 | 0.40 | 0.46 | 0.000000 |
| 2 | 0 – 120 | - | 6 | 120 | 72.08 | 0.13 | 0.43 | 0.000000 |
| 3 | 40 – 160 | + | 6 | 120 | 79.66 | -0.23 | 0.55 | 0.003969 |
| 4 | 40 – 160 | - | 6 | 120 | 79.66 | -0.97 | 0.73 | 0.474261 |
| 5 | 80 – 200 | + | 6 | 120 | 82.48 | -1.34 | 0.55 | 0.145823 |
| 7 | 120 – 240 | + | 6 | 120 | 86.68 | -0.59 | 0.71 | 0.113574 |
| 8 | 120 – 240 | - | 6 | 120 | 86.68 | -1.01 | 0.72 | 0.150117 |
| 9 | 160 – 280 | + | 6 | 120 | 86.11 | -0.48 | 0.76 | 0.074647 |
| 10 | 160 – 280 | - | 6 | 120 | 86.11 | -0.79 | 0.64 | 0.144259 |
| 11 | 200 – 320 | + | 6 | 120 | 85.49 | -0.20 | 0.71 | 0.084988 |
| 12 | 200 – 320 | - | 6 | 120 | 85.49 | -0.33 | 0.72 | 0.089235 |
| 13 | 240 – 360 | + | 6 | 120 | 84.15 | 1.32 | 0.55 | 0.008851 |
| 14 | 240 – 360 | - | 6 | 120 | 84.15 | 0.50 | 0.53 | 0.015067 |
| 15 | 280 – 400 | + | 6 | 120 | 74.88 | -0.24 | 0.26 | 0.000000 |
| 16 | 280 – 400 | - | 6 | 120 | 74.88 | 1.68 | 0.39 | 0.000000 |
| 17 | 320 – 440 | + | 6 | 120 | 80.61 | 0.69 | 0.56 | 0.000002 |
| 18 | 320 – 440 | - | 6 | 120 | 80.61 | 1.78 | 0.49 | 0.000000 |
| 19 | 360 – 480 | + | 2 | 120 | 54.78 | -0.42 | 0.47 | 0.000000 |
| 20 | 360 – 480 | - | 2 | 120 | 54.78 | 0.39 | 0.47 | 0.000000 |
| 21 | 440 – 560 | + | 2 | 120 | 83.19 | -0.36 | 0.76 | 0.055207 |
| 22 | 440 – 560 | - | 2 | 120 | 83.19 | 0.47 | 0.81 | 0.028617 |
| 23 | 520 – 640 | + | 6 | 120 | 74.60 | -0.18 | 0.45 | 0.000000 |
| 24 | 520 – 640 | - | 6 | 120 | 74.60 | -0.50 | 0.41 | 0.000000 |
| 25 | 560 – 680 | + | 6 | 120 | 81.30 | -0.52 | 0.48 | 0.004076 |
| 26 | 560 – 680 | - | 6 | 120 | 81.30 | -0.32 | 0.56 | 0.014577 |
| 27 | 600 – 720 | + | 6 | 120 | 80.66 | 0.22 | 0.47 | 0.000000 |
| 28 | 600 – 720 | - | 6 | 120 | 80.66 | 0.18 | 0.46 | 0.000000 |
| 29 | 640 – 760 | + | 6 | 120 | 78.42 | -0.28 | 0.39 | 0.000000 |
| 30 | 640 – 760 | - | 6 | 120 | 78.42 | 0.02 | 0.21 | 0.000000 |
| 31 | 680 – 800 | + | 6 | 120 | 84.91 | 0.19 | 0.60 | 0.072917 |
| 32 | 680 – 800 | - | 6 | 120 | 84.91 | -0.85 | 0.59 | 0.129229 |
| 33 | 720 – 840 | + | 6 | 120 | 91.88 | -0.69 | 0.78 | 0.027538 |
| 35 | 760 – 880 | + | 6 | 120 | 81.07 | -1.15 | 0.49 | 0.032073 |
| 36 | 760 – 880 | - | 6 | 120 | 81.07 | -1.22 | 0.52 | 0.064611 |
| 37 | 800 – 920 | + | 6 | 120 | 85.98 | -1.36 | 0.57 | 0.087232 |
| 38 | 800 – 920 | - | 6 | 120 | 85.98 | -0.84 | 0.53 | 0.055267 |
| 39 | 840 – 960 | + | 6 | 120 | 81.89 | -0.63 | 0.61 | 0.091427 |
| 40 | 840 – 960 | - | 6 | 120 | 81.89 | -1.27 | 0.68 | 0.466952 |
| 41 | 880 – 1000 | + | 6 | 120 | 76.47 | -0.44 | 0.49 | 0.000001 |
| 42 | 880 – 1000 | - | 6 | 120 | 76.47 | -0.84 | 0.59 | 0.063718 |
| 43 | 920 – 1040 | + | 6 | 120 | 73.77 | -0.25 | 0.51 | 0.000000 |
| 44 | 920 – 1040 | - | 6 | 120 | 73.77 | 0.14 | 0.46 | 0.000000 |
| 45 | 960 – 1080 | + | 6 | 120 | 81.08 | -0.26 | 0.67 | 0.062385 |
| 46 | 960 – 1080 | - | 6 | 120 | 81.08 | 0.31 | 0.66 | 0.012981 |
| 47 | 1000 – 1120 | + | 6 | 120 | 87.05 | -0.93 | 0.71 | 0.128093 |
| 48 | 1000 – 1120 | - | 6 | 120 | 87.05 | -0.78 | 0.54 | 0.061687 |
| 49 | 1040 – 1160 | + | 6 | 120 | 87.46 | -0.05 | 0.60 | 0.128939 |
| 50 | 1040 – 1160 | - | 6 | 120 | 87.46 | -0.16 | 0.71 | 0.068865 |
| 51 | 1080 – 1200 | + | 6 | 120 | 83.60 | -1.20 | 0.70 | 0.343656 |
| 52 | 1080 – 1200 | - | 6 | 120 | 83.60 | 0.01 | 0.56 | 0.032916 |
| 53 | 1096 – 1216 | + | 6 | 120 | 89.30 | -0.80 | 0.82 | 0.068678 |
| 54 | 1096 – 1216 | - | 6 | 120 | 89.30 | -0.12 | 0.82 | 0.010667 |