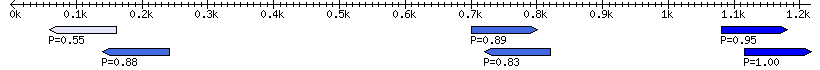
| Window 101 | |
|
| Window 97 | |
|
| Window 62 | |
|
| Window 59 | |
|
| Window 16 | |
|
| Window 8 | |
|
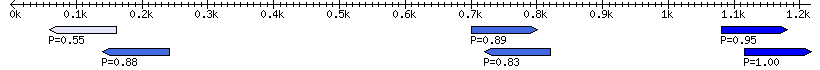
| Location | 60 – 160 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 82.11 |
| Mean single sequence MFE | -39.67 |
| Consensus MFE | -30.18 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.550349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
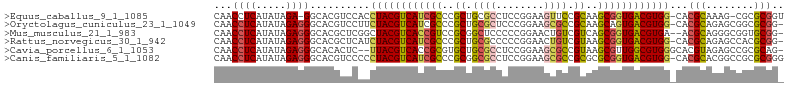
>Equus_caballus_9_1_1085/60-160 CAACCUCAUAUAGA-GGCACGUCCACCUACGUCAUCGCCCGCUGCGCCUCCGGAAGUUCCGCAAGCGGUGACGUGG-CACGCAAAG-CGCGCGGU ...((((.....))-)).......((((((((((((((....((((....(....)...)))).)))))))))))(-(.(((...)-)).))))) ( -34.60) >Oryctolagus_cuniculus_23_1_1049/60-160 CAACCUCAUAUAGAGGGCACGUCCUUCUACGUCAUCGCCCGCUGCGCUCCCGGAAGCGCCGCAAGCAGUGACGUGG-CACGCAGAGCGGCGCGG- ...((((.....))))((.((..((.(((((((((.((..((.(((((......))))).))..)).)))))))))-.....))..))))....- ( -37.60) >Mus_musculus_21_1_983/60-160 CAACCUCAUAUAGAGGGCACGCUCGGCUACGUCACCGUCCGCGGCUCCCCCGGAACUGUCGUCAGCGGUGACGUGA--ACGCAGGGCGGUGCGG- ...((((.....))))((((((((.(((((((((((((..((((((((...)))...)))))..))))))))))).--..)).)))).))))..- ( -45.40) >Rattus_norvegicus_30_1_942/60-160 CAACCUCAUAUAGAGGGCACGCUCAUCUACGUCAUCGCCCGCUGCGCCCCCGGAACUGUCGUAAGCGGUGACGUGG-CACGCAGAGCCACGCGG- ...((((.....))))...(((........((((((((..((.((.((...))....)).))..))))))))((((-(.......)))))))).- ( -33.40) >Cavia_porcellus_6_1_1053/60-160 CAACCUCAUAUAGAGGGCACACUC--UUACGUCACCGCGUGCUGCGCCUCCGGAAGCGCCGUAAGCGUUGGCGUGGGCACGUAGAGCCGCGCAG- ...((((.....))))((.(.(((--(.((((..((((((...((((...(((.....)))...))))..))))))..))))))))..).))..- ( -35.90) >Canis_familiaris_5_1_1082/60-160 CAACCUCAUAUAGAGGGCACGUCCCCCUACGUCAUCGCCCGCGGCGCCUCCGGAAGCGCCGCGCGCGGUGACGUGG-CACGCACGGCCGCGCGGG ...((((.....)))).......(((((((((((((((.((((((((........)))))))).))))))))))))-..(((......))).))) ( -51.10) >consensus CAACCUCAUAUAGAGGGCACGCCCACCUACGUCAUCGCCCGCUGCGCCCCCGGAAGCGCCGCAAGCGGUGACGUGG_CACGCAGAGCCGCGCGG_ ...((((.....))))((........((((((((((((..((.((((........)))).))..))))))))))))....((......))))... (-30.18 = -30.30 + 0.12)
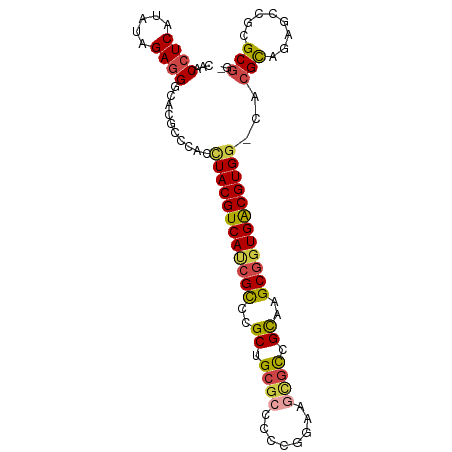
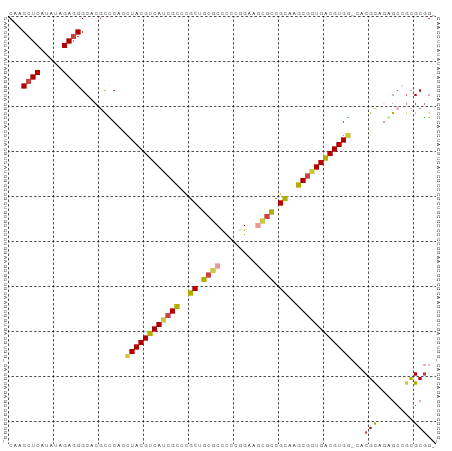
| Location | 140 – 240 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 92.07 |
| Mean single sequence MFE | -46.85 |
| Consensus MFE | -38.97 |
| Energy contribution | -39.67 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Pan_troglodytes_25_1_1076/140-240 AGCGAGCGCGGUGCAGCCGGAGCAGCGCUCGGGGGGAGGGGGGAGCGGGCGGAAAAGGCCGACUCAGCCGCUCCCCAACCUCAUAUAGAGGGCAC .((((((((..(((.......)))))))))..((((((.((.((((((.(......).))).)))..)).))))))..((((.....)))))).. ( -50.00) >Microcebus_murinus_19_1_1098/140-240 AGCGAGCGCGGUGGAGCCGGAGCAGCGCUCGGGGGGAGGGGGGAGCGGGCGGAAAAGGCCGAUUCAGCCGCUCCCCAACCUCAUAUAGAGGGCAC .((((((((.((.........)).))))))..((((((.((.((((((.(......).))).)))..)).))))))..((((.....)))))).. ( -46.30) >Equus_caballus_9_1_1085/140-240 ACCGAGCGGGGUGCUUCCG-AGC-GCGCUCAGGGG--AGGGGGAGCGGGCGGAAAA-GCCGACUCAGCCGCUCCCCAACCUCAUAUAGA-GGCAC .((....)).(((((((.(-((.-...))).((((--..(((((((((.(((....-.)))......)))))))))..)))).....))-))))) ( -44.10) >Oryctolagus_cuniculus_23_1_1049/140-240 AGCGAGCGCGGUGCAGCCGGAGCGGCCCUCGGGGGUAGGGGGGAGCGGGCGGAAAAGGCCGACUCAGCCGCUCCCCAACCUCAUAUAGAGGGCAC ..((..(.(((.....))))..))((((((.(((((...(((((((((.(((......)))......))))))))).))))).....)))))).. ( -49.70) >Rattus_norvegicus_30_1_942/140-240 AGCGAGCGCGGUGCAGCCGGAGCGGCGCUCGGGGGGAGGGGGGAGCGGGCGGAAAAGGCCGACUCGGGCGCUUCCCAACCUCAUAUAGAGGGCAC .((((((((.((.(.....).)).))))))..((((((.(..((((((.(......).))).)))...).))))))..((((.....)))))).. ( -43.10) >Cavia_porcellus_6_1_1053/140-240 GGCGAGCGCGGUGCAGCCGCAGCGGCACUCGGGGGGAGGGGGGAGCGGGCGGAAAAGGCCGACUCAGCCGCUCCCCAACCUCAUAUAGAGGGCAC ..((((.((.((((....))).).)).)))).((((((.((.((((((.(......).))).)))..)).))))))..((((.....)))).... ( -47.90) >consensus AGCGAGCGCGGUGCAGCCGGAGCGGCGCUCGGGGGGAGGGGGGAGCGGGCGGAAAAGGCCGACUCAGCCGCUCCCCAACCUCAUAUAGAGGGCAC .((((((((.((.........)).)))))).((((....(((((((((.(((......)))......)))))))))..)))).........)).. (-38.97 = -39.67 + 0.69)

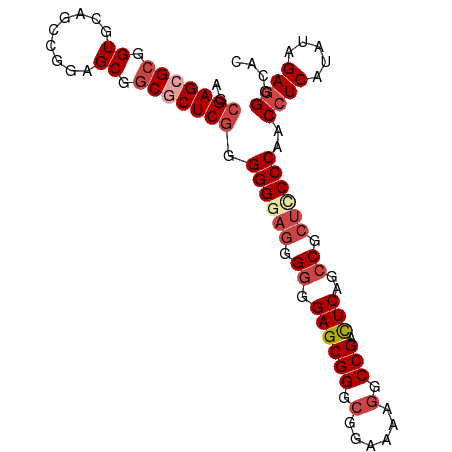
| Location | 700 – 800 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 79 |
| Reading direction | forward |
| Mean pairwise identity | 70.61 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -23.04 |
| Energy contribution | -24.60 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
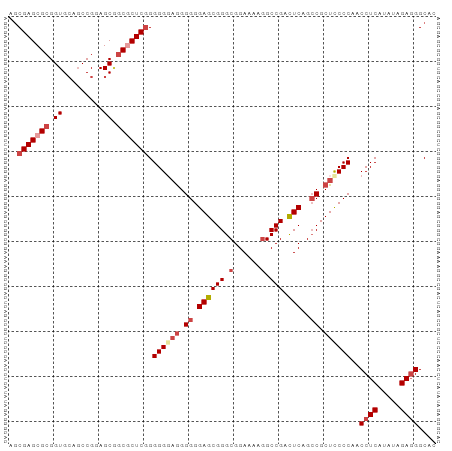
>Homo_sapiens_14_1_1076/700-800 GGGCCUGCGCGUGGGGGAGUCCGGG------CCGAGCGGGCUCGGGUUUCCUCCGCUCCCCCGCCUGAGGUUGUGCAAU .(((((.((.(((((((((((((((------((.....))))))).........)))))))))).)))))))....... ( -38.70) >Pan_troglodytes_25_1_1076/700-800 AGGCCUGCGCGUGGGGGAGUCCGGG------CCGAGCAGGCUCGGGUUUCCGCCGCUCCCCCGCCUGAGGUUGUGCAAU .(((((.((.((((((((((.((((------((.....))))))(((....))))))))))))).)))))))....... ( -40.10) >Pongo_pygmaeus_28_1_1078/700-800 GGGCCUGCGCGUGGGGGAGUCCGGG------CCGAGCAGGCUCGGGUUUCCGCCGCUCCCCCGCCUGAGGUUGUGCAAU .(((((.((.((((((((((.((((------((.....))))))(((....))))))))))))).)))))))....... ( -40.20) >Rattus_norvegicus_30_1_942/700-800 GUCG----UGGGGAGGUUGGCCGGUCGCCGGCCGG-CCGGGCUGAGCU--UGCGGGGAGGCCGCUGGAGCUCGUGCAAU ....----......((((((((((...))))))))-))..((((((((--(((((.....))))..))))))).))... ( -37.30) >consensus GGGCCUGCGCGUGGGGGAGUCCGGG______CCGAGCAGGCUCGGGUUUCCGCCGCUCCCCCGCCUGAGGUUGUGCAAU .(((((.((.(((((((((..((((......(((((....)))))......))))))))))))).)))))))....... (-23.04 = -24.60 + 1.56)
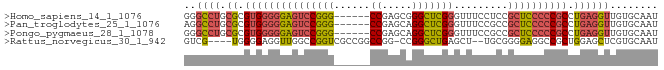
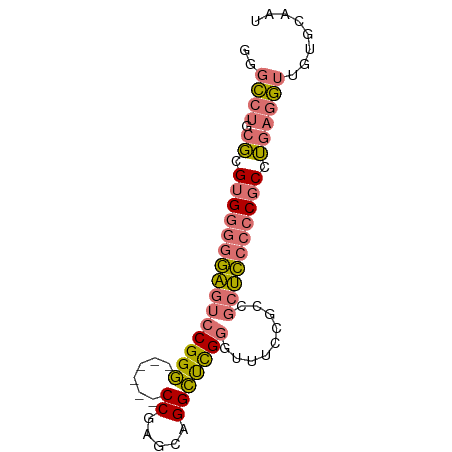
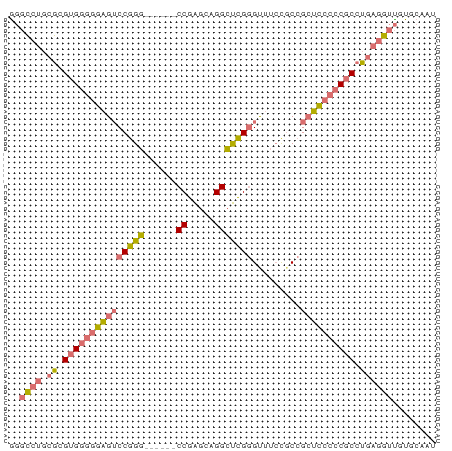
| Location | 720 – 820 |
|---|---|
| Length | 100 |
| Sequences | 2 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -34.80 |
| Consensus MFE | -34.55 |
| Energy contribution | -34.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.834102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Equus_caballus_9_1_1085/720-820 GCCAGGAGGCAGCGGAGAGGAUUGCACAACCUCGGGCGGGGGAGCGGCGGAAACCCGAGCCUGCCCGGCCUCG .....(((((......((((.((....))))))(((((((....(((.(....))))..))))))).))))). ( -36.50) >Felis_catus_10_1_1097/720-820 GCCAGGAGGCAGCGGAGAGGAUUGCACAACUUCGGGCGGGGGUGCGGCGGAAACCCGAGCCUGCCCGGCCUUG .....(((((.((((......)))).......((((((((....(((.(....))))..))))))))))))). ( -33.10) >consensus GCCAGGAGGCAGCGGAGAGGAUUGCACAACCUCGGGCGGGGGAGCGGCGGAAACCCGAGCCUGCCCGGCCUCG .....(((((......((((.((....))))))(((((((....(((.(....))))..))))))).))))). (-34.55 = -34.05 + -0.50)

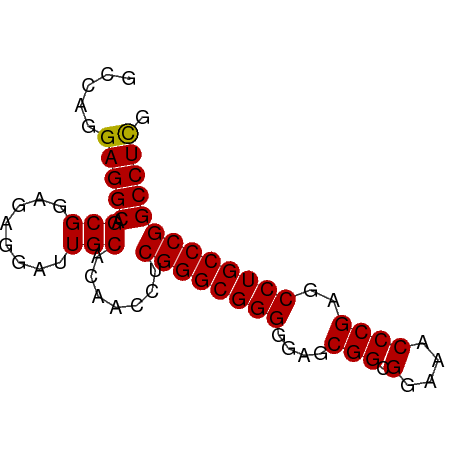
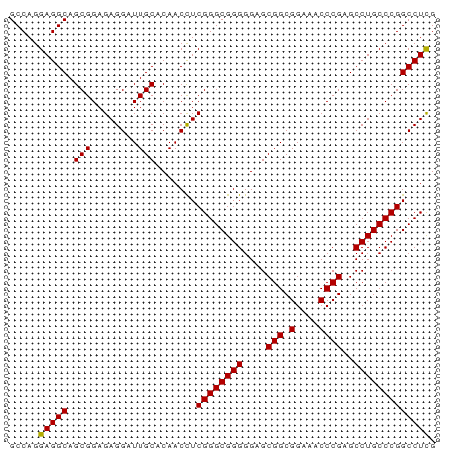
| Location | 1,080 – 1,180 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 91.09 |
| Mean single sequence MFE | -20.83 |
| Consensus MFE | -18.19 |
| Energy contribution | -17.25 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.948806 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Macaca_mulatta_17_1_1076/1080-1180 AGCACAGUAAUCACUGGUGUCGAUAUUGUCAUGAACCAUCACCUGCAGGAAACAAGUUUCACGAAAGAAGCCUACAAGAAGUACAUCAAAGAUUACAU ......((((((..(((((((....((((..................(....)..(((((......)))))..))))...).))))))..)))))).. ( -20.20) >Microcebus_murinus_19_1_1098/1080-1180 AGCACAGUAAUCACUGGUGUUGAUAUCGUCAUGAACCAUCAUUUGCAAGAAACCAGCUUCACAAAAGAAGCCUACAAGAAGUACAUCAAAGAUUACAU ......((((((..((((((.....((((.((((....))))..)).........(((((......)))))......))...))))))..)))))).. ( -19.90) >Mus_musculus_21_1_983/1080-1180 AGCACAGUAGUCACCGGUGUUGACAUUGUCAUGAACCAUCACUUACAAGAAACCAGCUUCACAAAAGAGGCUUACAAAAAGUACAUCAAAGACUACAU ......((((((...(((..((((...))))...)))......(((........((((((......))))))........))).......)))))).. ( -22.69) >Rattus_norvegicus_30_1_942/1080-1180 AGCACAGUAGUCACCGGUGUUGACAUUGUCAUGAACCAUCACUUACAAGAAACCAGCUUCACAAAAGAGGCCUACAAAAAGUAUAUCAAAGACUACAU ......((((((...(((..((((...))))...)))..................(((((......)))))...................)))))).. ( -21.60) >Cavia_porcellus_6_1_1053/1080-1180 AGCACAGUAAUCACUGGUGUUGAUAUUGUCAUGAACCAUCACUUGCAGGAAACCAGUUUCACAAAAGAGGCCUACAAGAAGUACAUCAAAGAUUAUAU ......((((((..((((..((((...))))...))))...((((..(....)..(((((......)))))...))))............)))))).. ( -17.40) >Felis_catus_10_1_1097/1080-1180 AGCACAGUAAUCACUGGUGUGGAUAUCGUCAUGAACCAUCACUUGCAGGAAACCAGCUUCACAAAGGAAGCCUACAAGAAGUACAUCAAAGAUUACAU ......((((((..(((((((..........(((....)))((((..(....)..(((((......)))))...))))...)))))))..)))))).. ( -23.20) >consensus AGCACAGUAAUCACUGGUGUUGAUAUUGUCAUGAACCAUCACUUGCAAGAAACCAGCUUCACAAAAGAAGCCUACAAGAAGUACAUCAAAGAUUACAU ......((((((...(((..((((...))))...)))..................(((((......)))))...................)))))).. (-18.19 = -17.25 + -0.94)


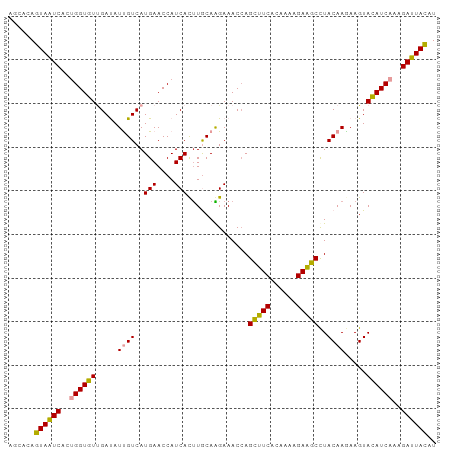
| Location | 1,116 – 1,216 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 90.06 |
| Mean single sequence MFE | -15.92 |
| Consensus MFE | -15.22 |
| Energy contribution | -15.13 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.96 |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999786 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Microcebus_murinus_19_1_1098/1116-1216 ACCAUCAUUUGCAAGAAACCAGCUUCACAAAAGAAGCCUACAAGAAGUACAUCAAAGAUUACAUGAAGUCGUAAGUA--- ......((((((.........(((((......)))))...................((((......)))))))))).--- ( -11.60) >Mus_musculus_21_1_983/1116-1216 ACCAUCACUUACAAGAAACCAGCUUCACAAAAGAGGCUUACAAAAAGUACAUCAAAGACUACAUGAAAUCGUAAGUGACA ....((((((((........((((((......))))))(((.....))).....................)))))))).. ( -17.10) >Rattus_norvegicus_30_1_942/1116-1216 ACCAUCACUUACAAGAAACCAGCUUCACAAAAGAGGCCUACAAAAAGUAUAUCAAAGACUACAUGAAAUCGUAAGUGACG ....((((((((.........(((((......))))).(((.....))).....................)))))))).. ( -16.10) >Cavia_porcellus_6_1_1053/1116-1216 ACCAUCACUUGCAGGAAACCAGUUUCACAAAAGAGGCCUACAAGAAGUACAUCAAAGAUUAUAUGAAAUCGUAAGUG--- .....(((((((.(....)..(((((......)))))...................((((......)))))))))))--- ( -13.20) >Canis_familiaris_5_1_1082/1116-1216 ACCAUCACUUGCAGGAAACCAGCUUCACAAAAGAAGCCUACAAGAAGUACAUCAAAGAUUACAUGAAAUCGUAAGUGGUC ...(((((((((.(....)..(((((......)))))...................((((......))))))))))))). ( -18.70) >Felis_catus_10_1_1097/1116-1216 ACCAUCACUUGCAGGAAACCAGCUUCACAAAGGAAGCCUACAAGAAGUACAUCAAAGAUUACAUGAAAUCGUAAGUGAAC ....((((((((.(....)..(((((......)))))...................((((......)))))))))))).. ( -18.80) >consensus ACCAUCACUUGCAAGAAACCAGCUUCACAAAAGAAGCCUACAAGAAGUACAUCAAAGAUUACAUGAAAUCGUAAGUGA__ ....((((((((.........(((((......)))))...................((((......)))))))))))).. (-15.22 = -15.13 + -0.08)

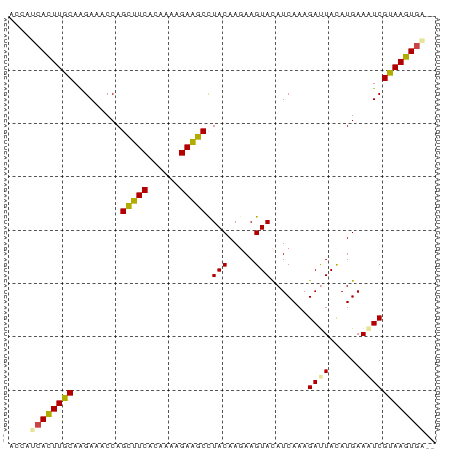
These windows have a probability below the cutoff.
| Window | Location | Strand | N | Length | Mean pairwise ID | z | SCI | P |
| 1 | 0 – 100 | + | 6 | 100 | 76.62 | 0.75 | 0.46 | 0.000000 |
| 2 | 0 – 100 | - | 6 | 100 | 76.62 | 0.56 | 0.50 | 0.000000 |
| 3 | 20 – 120 | + | 5 | 100 | 54.31 | -0.81 | 0.24 | 0.002836 |
| 4 | 20 – 120 | - | 5 | 100 | 54.31 | -1.14 | 0.29 | 0.051918 |
| 5 | 40 – 140 | + | 6 | 100 | 78.87 | -0.24 | 0.61 | 0.010620 |
| 6 | 40 – 140 | - | 6 | 100 | 78.87 | -0.16 | 0.71 | 0.044367 |
| 7 | 60 – 160 | + | 6 | 100 | 82.11 | -1.01 | 0.59 | 0.158480 |
| 9 | 80 – 180 | + | 6 | 100 | 85.02 | -0.85 | 0.48 | 0.023597 |
| 10 | 80 – 180 | - | 6 | 100 | 85.02 | -1.33 | 0.77 | 0.336243 |
| 11 | 100 – 200 | + | 6 | 100 | 87.59 | -0.85 | 0.69 | 0.107807 |
| 12 | 100 – 200 | - | 6 | 100 | 87.59 | -0.84 | 0.79 | 0.095466 |
| 13 | 120 – 220 | + | 6 | 100 | 87.59 | -0.77 | 0.69 | 0.106263 |
| 14 | 120 – 220 | - | 6 | 100 | 87.59 | -1.34 | 0.76 | 0.179914 |
| 15 | 140 – 240 | + | 6 | 100 | 92.07 | -1.50 | 0.84 | 0.458848 |
| 17 | 160 – 260 | + | 4 | 100 | 59.45 | -0.89 | 0.38 | 0.011875 |
| 18 | 160 – 260 | - | 4 | 100 | 59.45 | -0.61 | 0.43 | 0.017191 |
| 19 | 180 – 280 | + | 6 | 100 | 85.87 | -0.57 | 0.67 | 0.141851 |
| 20 | 180 – 280 | - | 6 | 100 | 85.87 | -0.56 | 0.64 | 0.142497 |
| 21 | 200 – 300 | + | 6 | 100 | 84.91 | 0.07 | 0.69 | 0.069570 |
| 22 | 200 – 300 | - | 6 | 100 | 84.91 | -0.00 | 0.67 | 0.085556 |
| 23 | 220 – 320 | + | 6 | 100 | 86.11 | 0.30 | 0.74 | 0.026692 |
| 24 | 220 – 320 | - | 6 | 100 | 86.11 | 0.27 | 0.73 | 0.034383 |
| 25 | 240 – 340 | + | 6 | 100 | 83.47 | 1.19 | 0.57 | 0.006726 |
| 26 | 240 – 340 | - | 6 | 100 | 83.47 | 0.78 | 0.56 | 0.009848 |
| 27 | 260 – 360 | + | 6 | 100 | 80.63 | 1.04 | 0.40 | 0.000000 |
| 28 | 260 – 360 | - | 6 | 100 | 80.63 | -0.01 | 0.54 | 0.002744 |
| 29 | 280 – 380 | + | 6 | 100 | 77.72 | 0.66 | 0.29 | 0.000000 |
| 30 | 280 – 380 | - | 6 | 100 | 77.72 | 1.31 | 0.38 | 0.000000 |
| 31 | 300 – 400 | + | 6 | 100 | 86.98 | -0.27 | 0.67 | 0.109552 |
| 32 | 300 – 400 | - | 6 | 100 | 86.98 | 1.45 | 0.63 | 0.042342 |
| 33 | 320 – 420 | + | 6 | 100 | 83.18 | -0.08 | 0.61 | 0.053573 |
| 34 | 320 – 420 | - | 6 | 100 | 83.18 | 1.43 | 0.52 | 0.000014 |
| 35 | 340 – 440 | + | 6 | 100 | 83.83 | 0.44 | 0.72 | 0.022652 |
| 36 | 340 – 440 | - | 6 | 100 | 83.83 | 1.16 | 0.35 | 0.000000 |
| 37 | 360 – 460 | + | 2 | 100 | 53.12 | -0.40 | 0.36 | 0.000000 |
| 38 | 360 – 460 | - | 2 | 100 | 53.12 | 0.58 | 0.32 | 0.000000 |
| 39 | 380 – 480 | + | 3 | 100 | 56.18 | -0.60 | 0.21 | 0.000000 |
| 40 | 380 – 480 | - | 3 | 100 | 56.18 | -0.16 | 0.21 | 0.000000 |
| 41 | 500 – 600 | + | 4 | 100 | 91.30 | -0.14 | 0.80 | 0.019092 |
| 42 | 500 – 600 | - | 4 | 100 | 91.30 | -0.10 | 0.79 | 0.018776 |
| 43 | 520 – 620 | + | 6 | 100 | 78.05 | -0.21 | 0.50 | 0.000000 |
| 44 | 520 – 620 | - | 6 | 100 | 78.05 | 0.00 | 0.41 | 0.000000 |
| 45 | 540 – 640 | + | 6 | 100 | 74.73 | -0.17 | 0.38 | 0.000000 |
| 46 | 540 – 640 | - | 6 | 100 | 74.73 | -0.07 | 0.49 | 0.000000 |
| 47 | 560 – 660 | + | 6 | 100 | 78.44 | 0.25 | 0.44 | 0.000000 |
| 48 | 560 – 660 | - | 6 | 100 | 78.44 | 0.14 | 0.54 | 0.000000 |
| 49 | 580 – 680 | + | 6 | 100 | 81.27 | 0.53 | 0.48 | 0.000000 |
| 50 | 580 – 680 | - | 6 | 100 | 81.27 | 0.03 | 0.50 | 0.000007 |
| 51 | 600 – 700 | + | 6 | 100 | 86.40 | 0.35 | 0.62 | 0.103226 |
| 52 | 600 – 700 | - | 6 | 100 | 86.40 | 0.33 | 0.57 | 0.098108 |
| 53 | 620 – 720 | + | 6 | 100 | 82.75 | -0.32 | 0.55 | 0.027319 |
| 54 | 620 – 720 | - | 6 | 100 | 82.75 | 0.17 | 0.55 | 0.011178 |
| 55 | 640 – 740 | + | 6 | 100 | 79.09 | -0.33 | 0.46 | 0.000001 |
| 56 | 640 – 740 | - | 6 | 100 | 79.09 | -0.71 | 0.24 | 0.000000 |
| 57 | 660 – 760 | + | 5 | 100 | 75.06 | 0.38 | 0.36 | 0.000000 |
| 58 | 660 – 760 | - | 5 | 100 | 75.06 | 0.16 | 0.23 | 0.000000 |
| 60 | 700 – 800 | - | 4 | 100 | 70.61 | -0.99 | 0.58 | 0.058343 |
| 61 | 720 – 820 | + | 2 | 100 | 95.89 | 0.27 | 0.95 | 0.015985 |
| 63 | 740 – 840 | + | 6 | 100 | 84.15 | -0.95 | 0.67 | 0.231923 |
| 64 | 740 – 840 | - | 6 | 100 | 84.15 | -1.38 | 0.65 | 0.292415 |
| 65 | 760 – 860 | + | 6 | 100 | 82.54 | -0.87 | 0.58 | 0.099388 |
| 66 | 760 – 860 | - | 6 | 100 | 82.54 | -1.42 | 0.51 | 0.084635 |
| 67 | 780 – 880 | + | 6 | 100 | 86.39 | -0.49 | 0.65 | 0.134175 |
| 68 | 780 – 880 | - | 6 | 100 | 86.39 | -0.38 | 0.61 | 0.128955 |
| 69 | 800 – 900 | + | 6 | 100 | 88.53 | -2.17 | 0.58 | 0.022207 |
| 70 | 800 – 900 | - | 6 | 100 | 88.53 | -0.97 | 0.59 | 0.056816 |
| 71 | 820 – 920 | + | 6 | 100 | 86.39 | -1.38 | 0.52 | 0.037077 |
| 72 | 820 – 920 | - | 6 | 100 | 86.39 | -0.93 | 0.38 | 0.000017 |
| 73 | 840 – 940 | + | 6 | 100 | 82.40 | -0.40 | 0.45 | 0.000018 |
| 74 | 840 – 940 | - | 6 | 100 | 82.40 | -1.10 | 0.49 | 0.026245 |
| 75 | 860 – 960 | + | 6 | 100 | 77.11 | -0.69 | 0.63 | 0.079966 |
| 76 | 860 – 960 | - | 6 | 100 | 77.11 | -1.13 | 0.59 | 0.205110 |
| 77 | 880 – 980 | + | 6 | 100 | 72.89 | -0.65 | 0.58 | 0.020256 |
| 78 | 880 – 980 | - | 6 | 100 | 72.89 | -0.81 | 0.62 | 0.141225 |
| 79 | 900 – 1000 | + | 6 | 100 | 78.91 | -0.72 | 0.47 | 0.002640 |
| 80 | 900 – 1000 | - | 6 | 100 | 78.91 | -0.94 | 0.59 | 0.116871 |
| 81 | 920 – 1020 | + | 6 | 100 | 76.69 | -0.26 | 0.43 | 0.000000 |
| 82 | 920 – 1020 | - | 6 | 100 | 76.69 | -0.28 | 0.60 | 0.005035 |
| 83 | 940 – 1040 | + | 6 | 100 | 72.00 | 0.00 | 0.51 | 0.000000 |
| 84 | 940 – 1040 | - | 6 | 100 | 72.00 | -0.03 | 0.13 | 0.000000 |
| 85 | 960 – 1060 | + | 6 | 100 | 78.80 | -0.21 | 0.69 | 0.046193 |
| 86 | 960 – 1060 | - | 6 | 100 | 78.80 | 0.07 | 0.49 | 0.000000 |
| 87 | 980 – 1080 | + | 6 | 100 | 89.83 | -0.70 | 0.82 | 0.048954 |
| 88 | 980 – 1080 | - | 6 | 100 | 89.83 | -0.33 | 0.61 | 0.072400 |
| 89 | 1000 – 1100 | + | 6 | 100 | 90.32 | -0.24 | 0.72 | 0.030052 |
| 90 | 1000 – 1100 | - | 6 | 100 | 90.32 | -0.31 | 0.62 | 0.061090 |
| 91 | 1020 – 1120 | + | 6 | 100 | 91.97 | -0.95 | 0.80 | 0.052992 |
| 92 | 1020 – 1120 | - | 6 | 100 | 91.97 | -0.58 | 0.69 | 0.019557 |
| 93 | 1040 – 1140 | + | 6 | 100 | 91.16 | 0.60 | 0.76 | 0.004156 |
| 94 | 1040 – 1140 | - | 6 | 100 | 91.16 | 0.08 | 0.87 | 0.002988 |
| 95 | 1060 – 1160 | + | 6 | 100 | 90.41 | -0.09 | 0.74 | 0.020928 |
| 96 | 1060 – 1160 | - | 6 | 100 | 90.41 | 0.79 | 0.77 | 0.003348 |
| 98 | 1080 – 1180 | - | 6 | 100 | 91.09 | -0.17 | 0.87 | 0.008136 |
| 99 | 1100 – 1200 | + | 6 | 100 | 89.07 | -1.08 | 0.81 | 0.137530 |
| 100 | 1100 – 1200 | - | 6 | 100 | 89.07 | -0.29 | 0.82 | 0.017564 |
| 102 | 1116 – 1216 | - | 6 | 100 | 90.06 | -0.93 | 0.54 | 0.022160 |