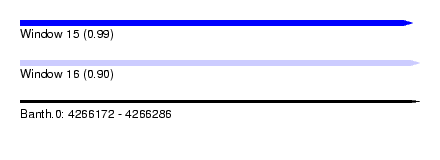
| Sequence ID | Banth.0 |
|---|---|
| Location | 4,266,172 – 4,266,286 |
| Length | 114 |
| Max. P | 0.988886 |

| Location | 4,266,172 – 4,266,284 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.00 |
| Mean single sequence MFE | -41.09 |
| Consensus MFE | -20.47 |
| Energy contribution | -18.84 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.93 |
| Mean z-score | -4.99 |
| Structure conservation index | 0.50 |
| SVM decision value | 2.14 |
| SVM RNA-class probability | 0.988886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4266172 112 + 5227293/0-120 GACGGAUUUGUCCGUUAUCAAGU-UAAGAGUAUAAGCAAAUUCCUGGAUUUGUUUGUAAAUAAAGGUGGUACCGCGA-UGUCCCUCGUCC-UUUUUU-----GGAUGAGGGACAUUUUUU ((((((....)))))).......-......((((((((((((....)))))))))))).......(((....)))((-((((((((((((-......-----)))))))))))))).... ( -45.50) >Bcere.0 4219445 113 + 5224283/0-120 GACGGAUUUGUCCGUUAUCAAAU-UAAGAGUAUAAACAAAUUCCUGAAUUUGUUUGUAAAUAAAGGUGGUACCGCGA-UGUCCCUCGUCCUUUUUUU-----GGAUGAGGGGCAUUUUUU ((((((....)))))).......-......((((((((((((....)))))))))))).......(((....)))((-((((((((((((.......-----)))))))))))))).... ( -43.50) >Bhalo.0 3155197 103 + 4202352/0-120 AGC---------CGUUAUCAUGU-CGAGUGUGCCAAUGG--UGAUGAUUCCAUAGGCAAACAAAGGUGGUACCGCGAGUAAACUCUCUUCGUCCUUU-----GAGCGAAGGGGGUUUUUU .((---------(((((((((.(-...((.((((.((((--........)))).)))).))..).))))))..))).))(((((((((((((.....-----..)))))))))))))... ( -34.90) >Bsubt.0 2868570 100 + 4214630/0-120 ACC---------CGUUAUCCGUUAUAAGUGCAUGAA-------CUGAUUGAGUUCAUGAAAAAAGGUGGUACCGCGAAAGAG--CUUUUCGUCCUUUUACAGGGAUGAAGAG--CUCUUU ...---------(((...(((.........((((((-------((.....))))))))........)))....)))((((((--(((((((((((......)))))))))))--)))))) ( -37.93) >Blich.0 2822565 110 + 4222334/0-120 UCC---------CGUUAUCCCAUUUAAGUGCGUGAAUGCG-UUUUUUUUGCAUUCAUGAAAAAAGGUGGUACCGCGAGAGAGUUCUCUUCGUCCUUUUAACAGGAUCAAGAGGACUCUUU ...---------(((....((((((.....((((((((((-.......)))))))))).....))))))....)))(((((((((((((.(((((......))))).))))))))))))) ( -43.60) >consensus AAC_________CGUUAUCAAAU_UAAGUGUAUAAACAA__UCCUGAUUGCGUUCGUAAAAAAAGGUGGUACCGCGA_UGACCCCUCUUCGUCCUUU_____GGAUGAAGGGCAUUUUUU ............(((...............((((((((((........))))))))))......((.....)))))...((((.((((((((((........)))))))))).))))... (-20.47 = -18.84 + -1.63)

| Location | 4,266,172 – 4,266,286 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.56 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -12.92 |
| Energy contribution | -10.92 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.87 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.899573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4266172 114 + 5227293/40-160 CAUCUUUUCU-GAACG---GAAUAGUUCUCAUUAGGAAAAGACGGAUUUGUCCGUUAUCAAGU-UAAGAGUAUAAGCAAAUUCCUGGAUUUGUUUGUAAAUAAAGGUGGUACCGCGA-UG ..((((((((-((((.---.....))))......))))))))((((....)))).((((....-......((((((((((((....)))))))))))).......(((....)))))-)) ( -28.70) >Bcere.0 4219445 114 + 5224283/40-160 CAUCUUUUCU-GAACG---GAAUAGUUCUCAUUAGGAAAAGACGGAUUUGUCCGUUAUCAAAU-UAAGAGUAUAAACAAAUUCCUGAAUUUGUUUGUAAAUAAAGGUGGUACCGCGA-UG ..((((((((-((((.---.....))))......))))))))((((....)))).((((....-......((((((((((((....)))))))))))).......(((....)))))-)) ( -27.80) >Bhalo.0 3155197 105 + 4202352/40-160 CAGCUUUACU-GAACA--CAAGUAAGUAAGGUCGGCGAAAAGC---------CGUUAUCAUGU-CGAGUGUGCCAAUGG--UGAUGAUUCCAUAGGCAAACAAAGGUGGUACCGCGAGUA ..((((((((-..((.--...)).))))))))((((.....))---------))........(-((.((.((((.((((--........)))).)))).))...((.....)).)))... ( -29.60) >Bsubt.0 2868570 103 + 4214630/40-160 CAUAAUUUCUUGAA-AGAAGAGUAGAGAAAUUCGGGAAACACC---------CGUUAUCCGUUAUAAGUGCAUGAA-------CUGAUUGAGUUCAUGAAAAAAGGUGGUACCGCGAAAG .....((((((...-.........))))))((((((...((((---------((.....)).........((((((-------((.....))))))))......))))...)).)))).. ( -22.30) >Blich.0 2822565 110 + 4222334/40-160 CGCAAUUUCUCGAACAAAAAAGUAGAGAAAUUCGGGAAAGUCC---------CGUUAUCCCAUUUAAGUGCGUGAAUGCG-UUUUUUUUGCAUUCAUGAAAAAAGGUGGUACCGCGAGAG (((((((((((..((......)).))))))))((((.....))---------)).....((((((.....((((((((((-.......)))))))))).....))))))....))).... ( -35.30) >consensus CAUCUUUUCU_GAACA___GAGUAGAUAAAAUCGGGAAAAAAC_________CGUUAUCAAAU_UAAGUGUAUAAACAA__UCCUGAUUGCGUUCGUAAAAAAAGGUGGUACCGCGA_UG ....................................................(((...............((((((((((........))))))))))......((.....))))).... (-12.92 = -10.92 + -2.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:15:02 2006