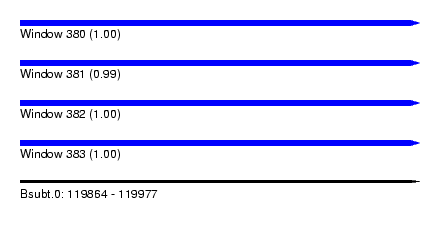
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 119,864 – 119,977 |
| Length | 113 |
| Max. P | 0.999745 |

| Location | 119,864 – 119,977 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.95 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -23.11 |
| Energy contribution | -22.18 |
| Covariance contribution | -0.94 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.66 |
| SVM decision value | 3.54 |
| SVM RNA-class probability | 0.999358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 119864 113 + 4214630/0-120 UAUACCGUAGACAGUAGGGGCCUA-ACCGCUUAAUUAUCCUACCGAGGU--GUAUAU-UAUCACAGCU--AUUACGUUACGUAUGCUUGUAUAUACAGCCUCCAUGUCU-CAUGGAGGCU ....(((((((((((((((((...-...))).......))))).(((((--((((((-...((.(((.--..((((...)))).))))).))))))).))))..)))))-.))))..... ( -33.11) >Blich.0 118834 114 + 4222334/0-120 UAUACCGUAGACAGUAGGUGCUGAUAAGGCUUAAUU-UCCUACCGAGGU--GUAUAUAUAUCACAGCU--CUAGCGUUACGUAUGCUUGUAUAUACAGCCUCCAUGUCG-AAUGGAGGCU ......(((.((((((((.(((.....)))......-.))))).(((.(--((.........))).))--).(((((.....))))))))...)))(((((((((....-.))))))))) ( -34.00) >Bhalo.0 151918 108 + 4202352/0-120 CAUACCGUAGACAGUAGGUGCGUGAUUCGCUUAAUU-UCCUACCGAGGUAAGCAAAUAGAUGAAAGUC--UU----UUGACUGGACUCGUAU-----GCCCUCAUGUCUGCAUGGGGGCA .((((........(((((.(((.....)))......-.))))).(((...((((((.((((....)))--))----))).))...)))))))-----(((((((((....))))))))). ( -38.30) >Bclau.0 167882 111 + 4303871/0-120 CAUACCGUAGACAGCAGGUGCUUC-AUAGCUUAAAUAUCCUGCCGAGGUGAGCGAAUAGAUCUCUUUUUACUAGAGCUUAAAGCCCUCGUGU-----GUCUUCGUGCC---ACGAGGGCU ...(((...(...(((((.(((..-..)))........))))))..)))((((...(((...........)))..))))..((((((((((.-----((......)))---))))))))) ( -34.20) >consensus CAUACCGUAGACAGUAGGUGCUUA_AUCGCUUAAUU_UCCUACCGAGGU__GCAAAUAGAUCACAGCU__CUAGCGUUAAGUAUGCUCGUAU_____GCCUCCAUGUCU_CAUGGAGGCU ..((((...(...(((((.(((.....)))........))))))..))))...............................................(((((((((....))))))))). (-23.11 = -22.18 + -0.94)

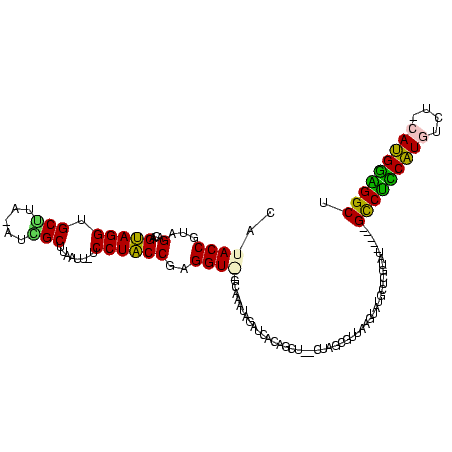
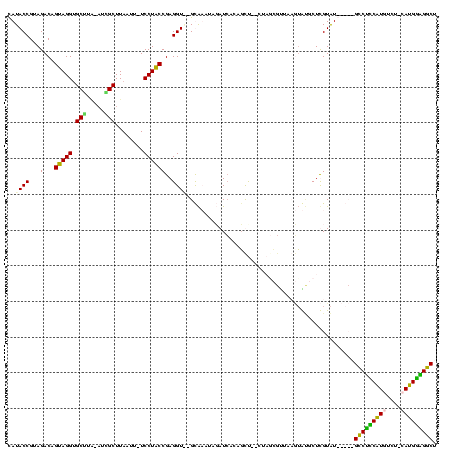
| Location | 119,864 – 119,977 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.95 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -19.75 |
| Energy contribution | -19.25 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.993593 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 119864 113 + 4214630/0-120 AGCCUCCAUG-AGACAUGGAGGCUGUAUAUACAAGCAUACGUAACGUAAU--AGCUGUGAUA-AUAUAC--ACCUCGGUAGGAUAAUUAAGCGGU-UAGGCCCCUACUGUCUACGGUAUA ((((((((((-...))))))))))(((((((((.((.((((...))))..--.)))))....-))))))--(((.(((((((.((((......))-))....))))))).....)))... ( -34.30) >Blich.0 118834 114 + 4222334/0-120 AGCCUCCAUU-CGACAUGGAGGCUGUAUAUACAAGCAUACGUAACGCUAG--AGCUGUGAUAUAUAUAC--ACCUCGGUAGGA-AAUUAAGCCUUAUCAGCACCUACUGUCUACGGUAUA (((((((((.-....)))))))))((((........)))).....(((.(--((.((((.......)))--).)))((((((.-........)))))))))...(((((....))))).. ( -30.90) >Bhalo.0 151918 108 + 4202352/0-120 UGCCCCCAUGCAGACAUGAGGGC-----AUACGAGUCCAGUCAA----AA--GACUUUCAUCUAUUUGCUUACCUCGGUAGGA-AAUUAAGCGAAUCACGCACCUACUGUCUACGGUAUG (((((.((((....)))).))))-----)...((....((((..----..--))))....))........((((.(((((((.-......(((.....))).))))))).....)))).. ( -28.80) >Bclau.0 167882 111 + 4303871/0-120 AGCCCUCGU---GGCACGAAGAC-----ACACGAGGGCUUUAAGCUCUAGUAAAAAGAGAUCUAUUCGCUCACCUCGGCAGGAUAUUUAAGCUAU-GAAGCACCUGCUGUCUACGGUAUG (((((((((---(..........-----.)))))))))).....((((.......))))............(((.(((((((...((((.....)-)))...))))))).....)))... ( -34.60) >consensus AGCCCCCAUG_AGACAUGAAGGC_____AUACAAGCACACGUAACGCUAG__AACUGUGAUAUAUAUAC__ACCUCGGUAGGA_AAUUAAGCGAU_UAAGCACCUACUGUCUACGGUAUA .((((((((......))))))))................................................(((.(((((((....................))))))).....)))... (-19.75 = -19.25 + -0.50)

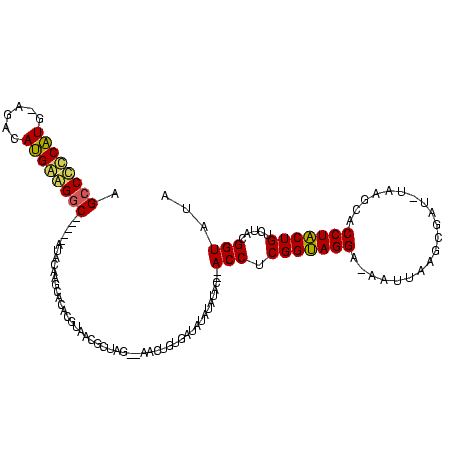
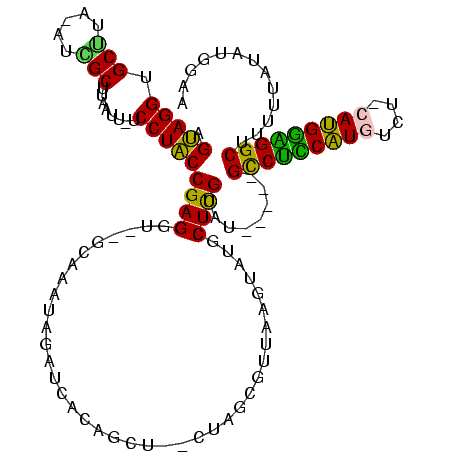
| Location | 119,864 – 119,977 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.67 |
| Mean single sequence MFE | -35.28 |
| Consensus MFE | -22.81 |
| Energy contribution | -21.18 |
| Covariance contribution | -1.62 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.65 |
| SVM decision value | 3.99 |
| SVM RNA-class probability | 0.999745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 119864 113 + 4214630/12-132 AGUAGGGGCCUA-ACCGCUUAAUUAUCCUACCGAGGU--GUAUAU-UAUCACAGCU--AUUACGUUACGUAUGCUUGUAUAUACAGCCUCCAUGUCU-CAUGGAGGCUUUUUAUAUGGAA .((((((((...-...))).......))))).....(--((((((-...((.(((.--..((((...)))).))))).)))))))(((((((((...-)))))))))............. ( -31.81) >Blich.0 118834 114 + 4222334/12-132 AGUAGGUGCUGAUAAGGCUUAAUU-UCCUACCGAGGU--GUAUAUAUAUCACAGCU--CUAGCGUUACGUAUGCUUGUAUAUACAGCCUCCAUGUCG-AAUGGAGGCUUUUUAUAUGGAA .(((((.(((.....)))......-.))))).(((.(--((.........))).))--)..((((.....))))(..((((...(((((((((....-.)))))))))...))))..).. ( -34.20) >Bhalo.0 151918 108 + 4202352/12-132 AGUAGGUGCGUGAUUCGCUUAAUU-UCCUACCGAGGUAAGCAAAUAGAUGAAAGUC--UU----UUGACUGGACUCGUAU-----GCCCUCAUGUCUGCAUGGGGGCAUUUUCGUUGGCC .(((((.(((.....)))......-.)))))((((...((((((.((((....)))--))----))).))...)))).((-----(((((((((....)))))))))))........... ( -39.30) >Bclau.0 167882 110 + 4303871/12-132 AGCAGGUGCUUC-AUAGCUUAAAUAUCCUGCCGAGGUGAGCGAAUAGAUCUCUUUUUACUAGAGCUUAAAGCCCUCGUGU-----GUCUUCGUGCC---ACGAGGGCUUUUC-UAUGGAA .(((((.(((..-..)))........))))).(((((..........)))))......(((.((...((((((((((((.-----((......)))---))))))))))).)-).))).. ( -35.80) >consensus AGUAGGUGCUUA_AUCGCUUAAUU_UCCUACCGAGGU__GCAAAUAGAUCACAGCU__CUAGCGUUAAGUAUGCUCGUAU_____GCCUCCAUGUCU_CAUGGAGGCUUUUUAUAUGGAA .(((((.(((.....)))........)))))((((......................................))))........(((((((((....)))))))))............. (-22.81 = -21.18 + -1.62)

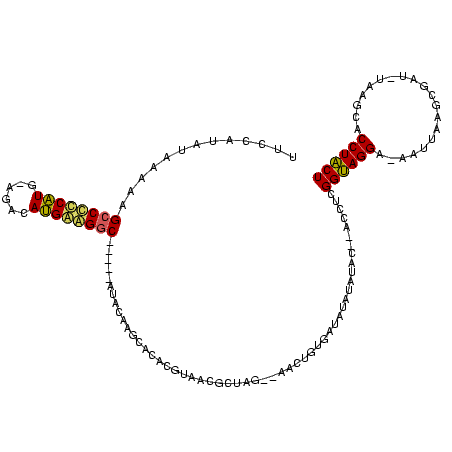
| Location | 119,864 – 119,977 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.67 |
| Mean single sequence MFE | -31.15 |
| Consensus MFE | -15.62 |
| Energy contribution | -15.12 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.50 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 119864 113 + 4214630/12-132 UUCCAUAUAAAAAGCCUCCAUG-AGACAUGGAGGCUGUAUAUACAAGCAUACGUAACGUAAU--AGCUGUGAUA-AUAUAC--ACCUCGGUAGGAUAAUUAAGCGGU-UAGGCCCCUACU ............((((((((((-...))))))))))(((((((((.((.((((...))))..--.)))))....-))))))--.....((((((.((((......))-))....)))))) ( -30.30) >Blich.0 118834 114 + 4222334/12-132 UUCCAUAUAAAAAGCCUCCAUU-CGACAUGGAGGCUGUAUAUACAAGCAUACGUAACGCUAG--AGCUGUGAUAUAUAUAC--ACCUCGGUAGGA-AAUUAAGCCUUAUCAGCACCUACU ....(((((...(((((((((.-....))))))))).)))))...............(((.(--((.((((.......)))--).)))((((((.-........)))))))))....... ( -29.60) >Bhalo.0 151918 108 + 4202352/12-132 GGCCAACGAAAAUGCCCCCAUGCAGACAUGAGGGC-----AUACGAGUCCAGUCAA----AA--GACUUUCAUCUAUUUGCUUACCUCGGUAGGA-AAUUAAGCGAAUCACGCACCUACU ((((.((....((((((.((((....)))).))))-----)).((((...((.(((----((--((......))).))))))...)))))).)).-......(((.....))).)).... ( -28.90) >Bclau.0 167882 110 + 4303871/12-132 UUCCAUA-GAAAAGCCCUCGU---GGCACGAAGAC-----ACACGAGGGCUUUAAGCUCUAGUAAAAAGAGAUCUAUUCGCUCACCUCGGCAGGAUAUUUAAGCUAU-GAAGCACCUGCU ....(((-(((((((((((((---(..........-----.))))))))))))...((((.......)))).)))))...........((((((...((((.....)-)))...)))))) ( -35.80) >consensus UUCCAUAUAAAAAGCCCCCAUG_AGACAUGAAGGC_____AUACAAGCACACGUAACGCUAG__AACUGUGAUAUAUAUAC__ACCUCGGUAGGA_AAUUAAGCGAU_UAAGCACCUACU .............((((((((......)))))))).....................................................((((((....................)))))) (-15.62 = -15.12 + -0.50)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:20:40 2006