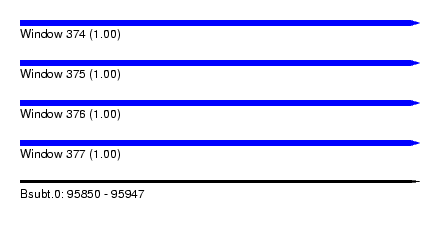
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 95,850 – 95,947 |
| Length | 97 |
| Max. P | 0.999710 |

| Location | 95,850 – 95,947 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -49.72 |
| Consensus MFE | -41.12 |
| Energy contribution | -44.68 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.36 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
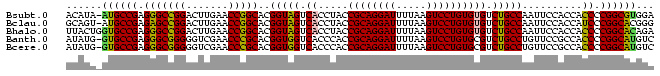
>Bsubt.0 95850 97 + 4214630 UCCACGCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCAAGUCCGGCCCUCGGCAU-UAUGU .....((((((((((..(((((((((......(((((.......)))))((((((....)))))))))))))))....)).)))).))))..-..... ( -47.60) >Bclau.0 142410 97 + 4303871 CCCGUGCCGGGAUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCAAGUCCGGCUCUCGGCAU-ACUGC ...((((((((((((..(((((((((......(((((.......)))))((((((....)))))))))))))))....)))..)))))))))-..... ( -47.00) >Bhalo.0 123861 98 + 4202352 UCUGUGCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCAAGUCCGGCCCUCGGCACCAGUAA .((((((((((((((..(((((((((......(((((.......)))))((((((....)))))))))))))))....)).)))).))))).)))... ( -51.00) >Banth.0 746830 97 + 5227293 GACAUGCCGGGGUGGCGGAACAGGCAGACGCACAGGACUUAAAAUCCUGCGGUGGGUGACCACCGUGCGGGUUCGACCCCCGCCCUCGGCAC-CAUAU ....(((((((((((.((..(..((...(((((((((.......))))..(((((....)))))))))).))..)..)))))))).))))).-..... ( -51.50) >Bcere.0 823523 97 + 5224283 GACAUGCCGGGGUGGCGGAACAGGCAGACGCACAGGACUUAAAAUCCUGCGGUGGGUGACCACCGUGCGGGUUCGACCCCCGCCCUCGGCAC-CAUAU ....(((((((((((.((..(..((...(((((((((.......))))..(((((....)))))))))).))..)..)))))))).))))).-..... ( -51.50) >consensus GCCAUGCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCAAGUCCGGCCCUCGGCAC_AAUAU ...((((((((((((..(((((((((......(((((.......)))))((((((....)))))))))))))))....)).)))).))))))...... (-41.12 = -44.68 + 3.56)

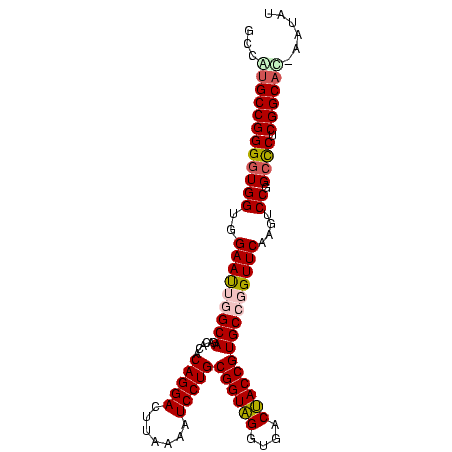
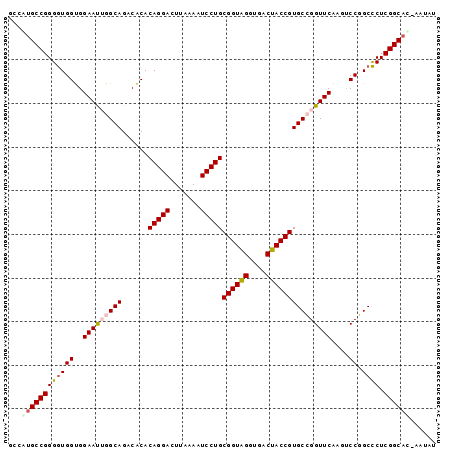
| Location | 95,850 – 95,947 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -41.02 |
| Energy contribution | -40.18 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 95850 97 + 4214630 ACAUA-AUGCCGAGGGCCGGACUUGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGCGUGGA .....-((((((.(((((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..........)).))))))... ( -42.10) >Bclau.0 142410 97 + 4303871 GCAGU-AUGCCGAGAGCCGGACUUGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCAUCCCGGCACGGG .....-.(((((.(((((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..........)).))))).... ( -39.30) >Bhalo.0 123861 98 + 4202352 UUACUGGUGCCGAGGGCCGGACUUGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGCACAGA ...(((.(((((.(((((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..........)).)))))))). ( -45.10) >Banth.0 746830 97 + 5227293 AUAUG-GUGCCGAGGGCGGGGGUCGAACCCGCACGGUGGUCACCCACCGCAGGAUUUUAAGUCCUGUGCGUCUGCCUGUUCCGCCACCCCGGCAUGUC .....-((((((..(((((((((.((...(((..(((((....)))))(((((((.....)))))))))))).))))...)))))....))))))... ( -46.00) >Bcere.0 823523 97 + 5224283 AUAUG-GUGCCGAGGGCGGGGGUCGAACCCGCACGGUGGUCACCCACCGCAGGAUUUUAAGUCCUGUGCGUCUGCCUGUUCCGCCACCCCGGCAUGUC .....-((((((..(((((((((.((...(((..(((((....)))))(((((((.....)))))))))))).))))...)))))....))))))... ( -46.00) >consensus AUAUG_GUGCCGAGGGCCGGACUUGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGCAUGGA ......((((((.(((((((.......)))))..(((((.((.....((((((((.....)))))))))).)))))..........)).))))))... (-41.02 = -40.18 + -0.84)
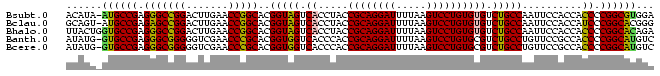
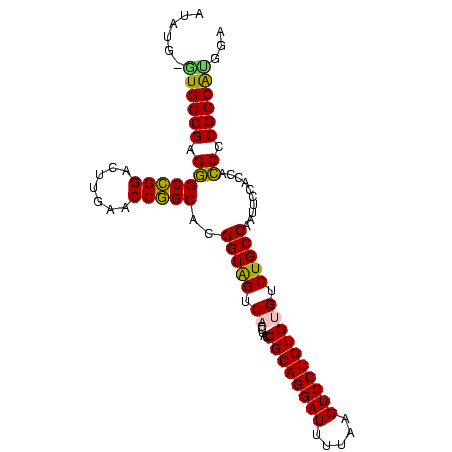
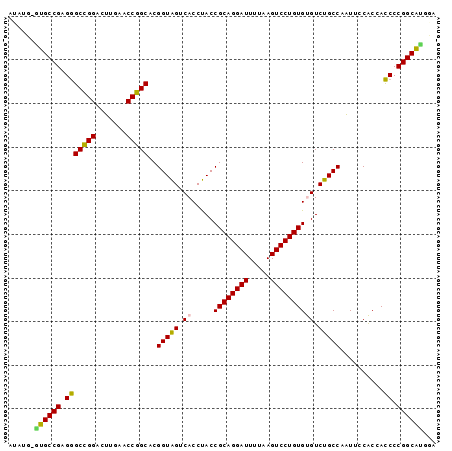
| Location | 95,850 – 95,947 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -49.72 |
| Consensus MFE | -41.12 |
| Energy contribution | -44.68 |
| Covariance contribution | 3.56 |
| Combinations/Pair | 1.16 |
| Mean z-score | -4.36 |
| Structure conservation index | 0.83 |
| SVM decision value | 3.76 |
| SVM RNA-class probability | 0.999592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 95850 97 + 4214630/0-98 UCCACGCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCAAGUCCGGCCCUCGGCAU-UAUGU .....((((((((((..(((((((((......(((((.......)))))((((((....)))))))))))))))....)).)))).))))..-..... ( -47.60) >Bclau.0 142410 97 + 4303871/0-98 CCCGUGCCGGGAUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCAAGUCCGGCUCUCGGCAU-ACUGC ...((((((((((((..(((((((((......(((((.......)))))((((((....)))))))))))))))....)))..)))))))))-..... ( -47.00) >Bhalo.0 123861 98 + 4202352/0-98 UCUGUGCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCAAGUCCGGCCCUCGGCACCAGUAA .((((((((((((((..(((((((((......(((((.......)))))((((((....)))))))))))))))....)).)))).))))).)))... ( -51.00) >Banth.0 746830 97 + 5227293/0-98 GACAUGCCGGGGUGGCGGAACAGGCAGACGCACAGGACUUAAAAUCCUGCGGUGGGUGACCACCGUGCGGGUUCGACCCCCGCCCUCGGCAC-CAUAU ....(((((((((((.((..(..((...(((((((((.......))))..(((((....)))))))))).))..)..)))))))).))))).-..... ( -51.50) >Bcere.0 823523 97 + 5224283/0-98 GACAUGCCGGGGUGGCGGAACAGGCAGACGCACAGGACUUAAAAUCCUGCGGUGGGUGACCACCGUGCGGGUUCGACCCCCGCCCUCGGCAC-CAUAU ....(((((((((((.((..(..((...(((((((((.......))))..(((((....)))))))))).))..)..)))))))).))))).-..... ( -51.50) >consensus GCCAUGCCGGGGUGGUGGAAUUGGCAGACACACAGGACUUAAAAUCCUGCGGUAGGUGACUACCGUGCCGGUUCAAGUCCGGCCCUCGGCAC_AAUAU ...((((((((((((..(((((((((......(((((.......)))))((((((....)))))))))))))))....)).)))).))))))...... (-41.12 = -44.68 + 3.56)

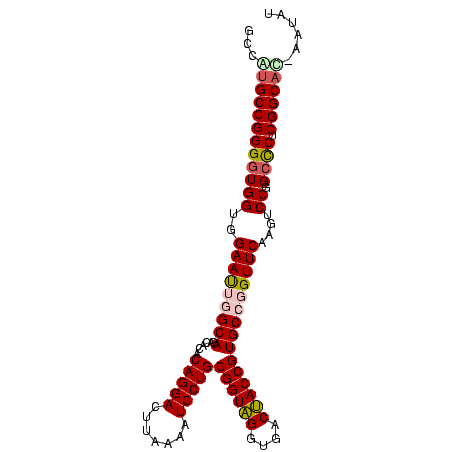
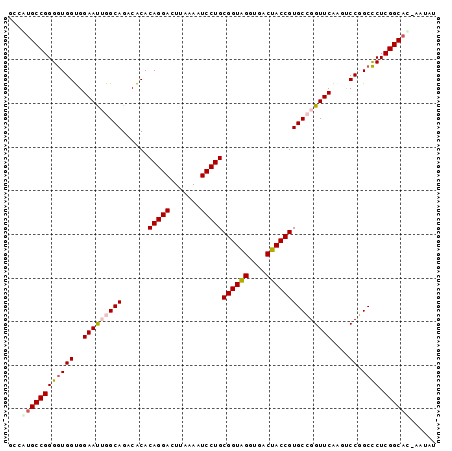
| Location | 95,850 – 95,947 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 85.63 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -41.02 |
| Energy contribution | -40.18 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 95850 97 + 4214630/0-98 ACAUA-AUGCCGAGGGCCGGACUUGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGCGUGGA .....-((((((.(((((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..........)).))))))... ( -42.10) >Bclau.0 142410 97 + 4303871/0-98 GCAGU-AUGCCGAGAGCCGGACUUGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCAUCCCGGCACGGG .....-.(((((.(((((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..........)).))))).... ( -39.30) >Bhalo.0 123861 98 + 4202352/0-98 UUACUGGUGCCGAGGGCCGGACUUGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGCACAGA ...(((.(((((.(((((((.......)))))..(((((.(((.....(((((((.....)))))))))).)))))..........)).)))))))). ( -45.10) >Banth.0 746830 97 + 5227293/0-98 AUAUG-GUGCCGAGGGCGGGGGUCGAACCCGCACGGUGGUCACCCACCGCAGGAUUUUAAGUCCUGUGCGUCUGCCUGUUCCGCCACCCCGGCAUGUC .....-((((((..(((((((((.((...(((..(((((....)))))(((((((.....)))))))))))).))))...)))))....))))))... ( -46.00) >Bcere.0 823523 97 + 5224283/0-98 AUAUG-GUGCCGAGGGCGGGGGUCGAACCCGCACGGUGGUCACCCACCGCAGGAUUUUAAGUCCUGUGCGUCUGCCUGUUCCGCCACCCCGGCAUGUC .....-((((((..(((((((((.((...(((..(((((....)))))(((((((.....)))))))))))).))))...)))))....))))))... ( -46.00) >consensus AUAUG_GUGCCGAGGGCCGGACUUGAACCGGCACGGUAGUCACCUACCGCAGGAUUUUAAGUCCUGUGUGUCUGCCAAUUCCACCACCCCGGCAUGGA ......((((((.(((((((.......)))))..(((((.((.....((((((((.....)))))))))).)))))..........)).))))))... (-41.02 = -40.18 + -0.84)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:20:36 2006