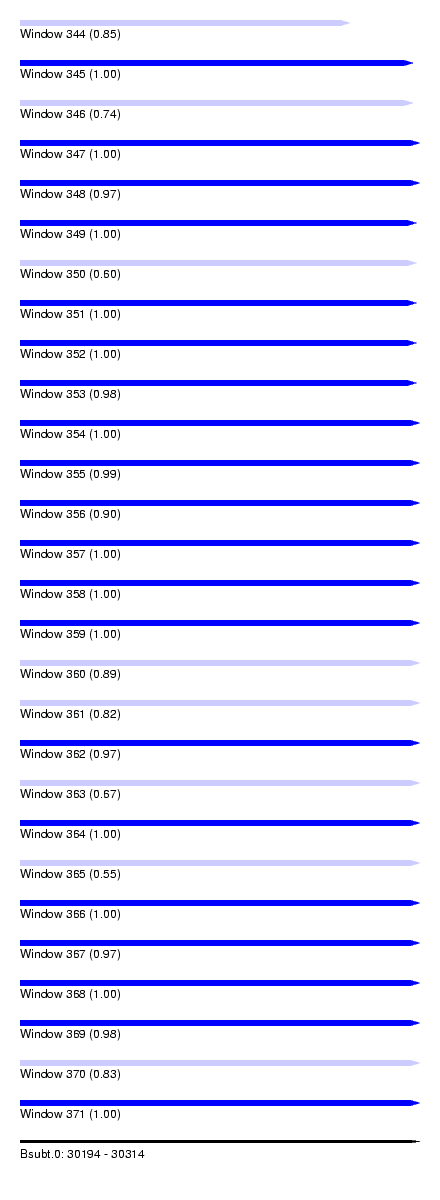
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 30,194 – 30,314 |
| Length | 120 |
| Max. P | 0.999998 |

| Location | 30,194 – 30,293 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.84 |
| Mean single sequence MFE | -33.36 |
| Consensus MFE | -28.04 |
| Energy contribution | -26.46 |
| Covariance contribution | -1.58 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 99 + 4214630/120-240 ----------UCAAACUAC---------UUUAUCGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGGACAGAU-GGGAGCUUGCUCCC- ----------(((((((.(---------((.....))))))))))(((((...))))).(..(((((((.(((((.......)))))..))).))))..)....-(((((....)))))- ( -34.60) >Blich.0 34457 118 + 4222334/120-240 UGUGACGAACUUAAUCUGCGCUCCAUUAUUUCAUGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGGACCGAU-GGGAGCUUGCUCCC- ..............(((((.((((((......)))))).(((((.(((((...))))))))))..((((.(((((.......)))))..)))).))))).....-(((((....)))))- ( -39.30) >Bcere.0 29012 99 + 5224283/120-240 ------------AACUAAC---------UUUAUUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAAUGGAUUAAGAGCUUGCUCUUA ------------..(.(((---------(((.....)))))).)((((((...))))))...(((((((.(((((.......)))))..))).)))).......((((((....)))))) ( -27.40) >Banth.0 29033 99 + 5227293/120-240 ------------AACUAAC---------UUUAUUGGAGAGUUUGAUCCUGGCUCAGGAUGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGAAUGGAUUAAGAGCUUGCUCUUA ------------..(.(((---------(((.....)))))).)((((((...))))))...(((((((.(((((.......)))))..))).)))).......((((((....)))))) ( -27.40) >Bclau.0 52263 117 + 4303871/120-240 -GUUGUGCAAUGAGCAAGU--CAAACUUUUUAACGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGGAGUUUCAAGAAGCUUGCUUUUU -.((((.(..(((((..((--(((((((((.....)))))))))))....)))))).)))).(((((((.(((((.......)))))..))).))))((((((....))))))....... ( -38.10) >consensus __________U_AACCAAC_________UUUAUUGGAGAGUUUGAUCCUGGCUCAGGACGAACGCUGGCGGCGUGCCUAAUACAUGCAAGUCGAGCGGACGGAU_AGGAGCUUGCUCUU_ .......................................(((((.(((((...))))))))))((((((.(((((.......)))))..))).)))........((((((....)))))) (-28.04 = -26.46 + -1.58)

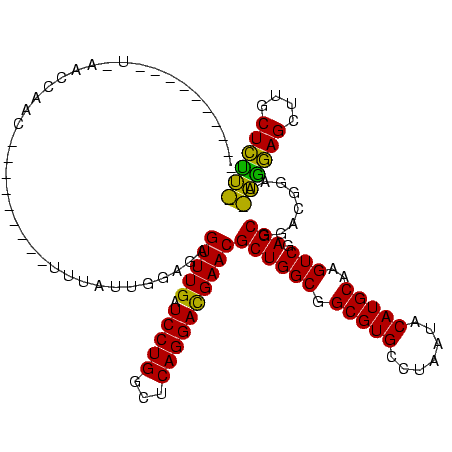
| Location | 30,194 – 30,312 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -44.82 |
| Consensus MFE | -44.58 |
| Energy contribution | -42.36 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.02 |
| SVM RNA-class probability | 0.999760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 118 + 4214630/200-320 UACAUGCAAGUCGAGCGGACAGAU-GGGAGCUUGCUCCC-UGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.((((...-(((((....)))))-...)))).)).....((((((..((.((......)).))..))))))..)))).).))..((((((....)))))).... ( -46.00) >Blich.0 34457 118 + 4222334/200-320 UACAUGCAAGUCGAGCGGACCGAU-GGGAGCUUGCUCCC-UUAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.((((...-(((((....)))))-...)))).)).....((((((..((.((......)).))..))))))..)))).).))..((((((....)))))).... ( -49.70) >Bcere.0 29012 120 + 5224283/200-320 UACAUGCAAGUCGAGCGAAUGGAUUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.(((....((((((....))))))....))).))(((((.((.(((.....))).))......))))).....)))).).))..((((((....)))))).... ( -38.90) >Banth.0 29033 120 + 5227293/200-320 UACAUGCAAGUCGAGCGAAUGGAUUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.(((....((((((....))))))....))).))(((((.((.(((.....))).))......))))).....)))).).))..((((((....)))))).... ( -38.90) >Bclau.0 52263 120 + 4303871/200-320 UACAUGCAAGUCGAGCGGAGUUUCAAGAAGCUUGCUUUUUGAAACUUAGCGGCGGACGGGUGAGUAACACGUGGGCAACCUGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAA ...((.(.((((..((.(((((((((((((....))))))))))))).))(((((.((.(((.....))).))(....)))))).....)))).).))..((((((....)))))).... ( -50.60) >consensus UACAUGCAAGUCGAGCGGACGGAU_AGGAGCUUGCUCUU_UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAA ...((.(.((((..((.((((...((((((....))))))...)))).))(((((.((.(((.....))).))(....)))))).....)))).).))..((((((....)))))).... (-44.58 = -42.36 + -2.22)

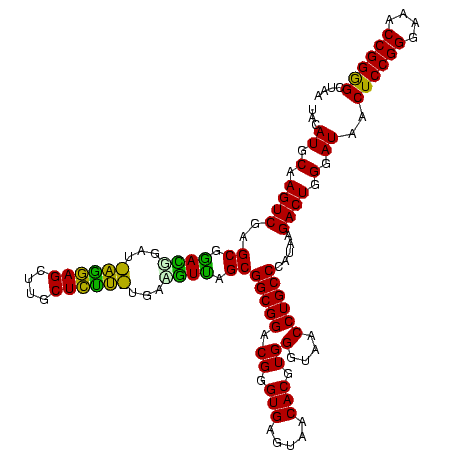
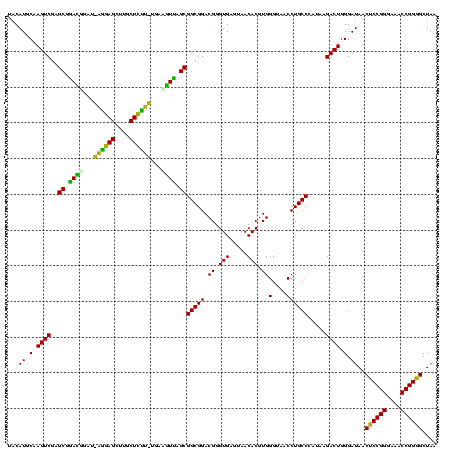
| Location | 30,194 – 30,312 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.73 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -30.12 |
| Energy contribution | -29.78 |
| Covariance contribution | -0.34 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.742951 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 118 + 4214630/200-320 UUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACAUCA-GGGAGCAAGCUCCC-AUCUGUCCGCUCGACUUGCAUGUA .((((.((((....)))).)))).........((((.(((((((......(((.....))).........((..(((...-(((((....)))))-...)))..))..))))))).)))) ( -34.70) >Blich.0 34457 118 + 4222334/200-320 UUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUGACCUAA-GGGAGCAAGCUCCC-AUCGGUCCGCUCGACUUGCAUGUA .((((.((((....)))).)))).........((((.(((((((......(((.....))).........((.((((...-(((((....)))))-...)))).))..))))))).)))) ( -40.20) >Bcere.0 29012 120 + 5224283/200-320 UUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACUUCAUAAGAGCAAGCUCUUAAUCCAUUCGCUCGACUUGCAUGUA .((((.((((....)))).))))..(((......)))(((((((......(((.....))).........((........((((((....))))))........))..)))))))..... ( -28.89) >Banth.0 29033 120 + 5227293/200-320 UUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACUUCAUAAGAGCAAGCUCUUAAUCCAUUCGCUCGACUUGCAUGUA .((((.((((....)))).))))..(((......)))(((((((......(((.....))).........((........((((((....))))))........))..)))))))..... ( -28.89) >Bclau.0 52263 120 + 4303871/200-320 UUAGCUCCGGUUUCCCGGAGUUAUCCCAGUCUAAGGGGCAGGUUGCCCACGUGUUACUCACCCGUCCGCCGCUAAGUUUCAAAAAGCAAGCUUCUUGAAACUCCGCUCGACUUGCAUGUA .(((((((((....))))))))).(((.......)))(((((((((..(((.((.....)).)))..)).((..((((((((.(((....))).))))))))..))..)))))))..... ( -42.20) >consensus UUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACGGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACUUCA_AAGAGCAAGCUCCU_AUCCAUCCGCUCGACUUGCAUGUA .((((.((((....)))).))))....((((.....(((.((....))..(((.....)))......)))((.((((...((((((....))))))...)))).))..))))........ (-30.12 = -29.78 + -0.34)

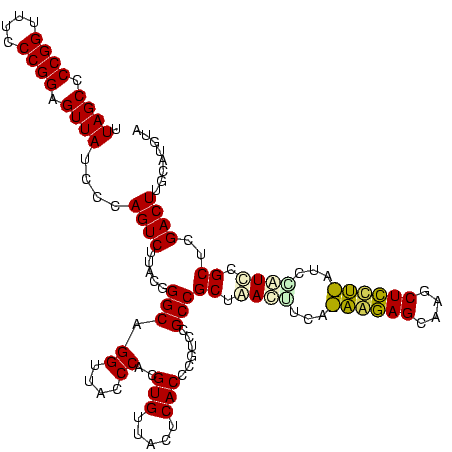
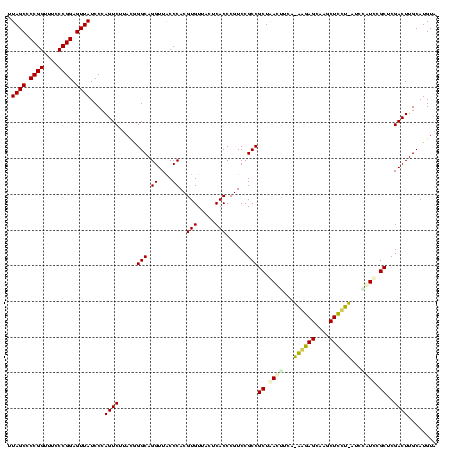
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -44.80 |
| Consensus MFE | -45.48 |
| Energy contribution | -42.28 |
| Covariance contribution | -3.20 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.13 |
| Structure conservation index | 1.02 |
| SVM decision value | 4.86 |
| SVM RNA-class probability | 0.999957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/240-360 UGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAACAUAAAA .(.((((((((((..((((((..((.((......)).))..))))))..).)))......((((((....)))))))))))).).......((((((((((....))))))))))..... ( -43.80) >Blich.0 34457 120 + 4222334/240-360 UUAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCAAUCAUAAAA .......((((.(..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).))))((((((((((....))))))))))..... ( -46.20) >Bcere.0 29012 120 + 5224283/240-360 UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAA ....((((.(((....((((((.....)))((((((.....))))))....)))......((((((....))))))......)))..))))((((((((((....))))))))))..... ( -41.80) >Banth.0 29033 120 + 5227293/240-360 UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCGAAAUUGAAA ....((((.((.(.....).))..))))..((((((.....))))))....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))..... ( -39.90) >Bclau.0 52263 120 + 4303871/240-360 GAAACUUAGCGGCGGACGGGUGAGUAACACGUGGGCAACCUGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAAUACCGGAUAAUCCCUUUCUCCACCUGGAGAGAGGGUGAAA ....((.((.(((((.((.(((.....))).))(....)))))).)).)).((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))..... ( -52.30) >consensus UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGUUUGAACCGCAUGGUUCGAAAAUGAAA ..........(((((.((.(((.....))).))(....)))))).......((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))..... (-45.48 = -42.28 + -3.20)

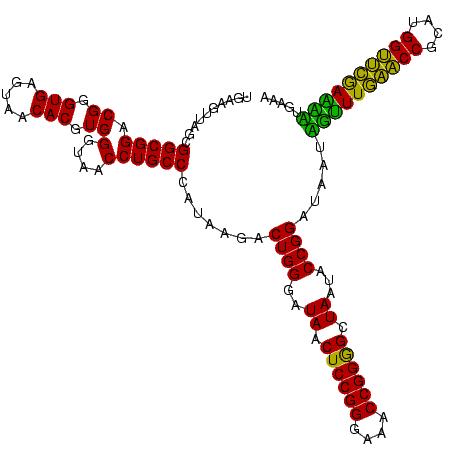
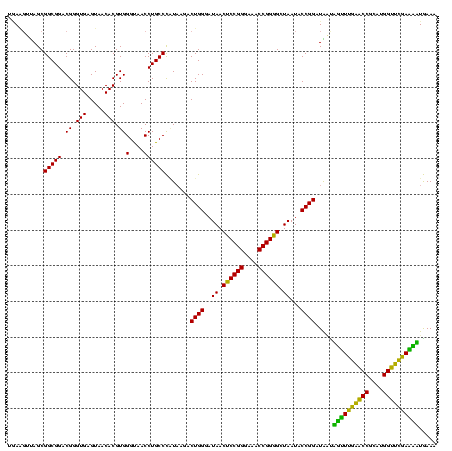
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.75 |
| Mean single sequence MFE | -34.86 |
| Consensus MFE | -31.72 |
| Energy contribution | -29.32 |
| Covariance contribution | -2.40 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968495 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/240-360 UUUUAUGUUUGAACCAUGCGGUUCAAACAACCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACAUCA .....((((((((((....)))))))))).......(((.(((((.((((....))))..................(((.((....))..(((.....)))......)))))))).))). ( -34.60) >Blich.0 34457 120 + 4222334/240-360 UUUUAUGAUUGAACCAUGCGGUUCAAUCAAGCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUGACCUAA .....((((((((((....))))))))))(((....((...((((.((((....)))).))))..)).........(((.((....))..(((.....)))......))))))....... ( -36.70) >Bcere.0 29012 120 + 5224283/240-360 UUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACUUCA .....((((.(((((....))))).))))((((..((((..((((.((((....)))).))))............((((.((....))..(((.....)))..)))))))).)))).... ( -31.10) >Banth.0 29033 120 + 5227293/240-360 UUUCAAUUUCGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACUUCA ..........(((....(((((..............((...((((.((((....)))).))))..))........((((.((....))..(((.....)))..)))))))))....))). ( -29.70) >Bclau.0 52263 120 + 4303871/240-360 UUUCACCCUCUCUCCAGGUGGAGAAAGGGAUUAUCCGGUAUUAGCUCCGGUUUCCCGGAGUUAUCCCAGUCUAAGGGGCAGGUUGCCCACGUGUUACUCACCCGUCCGCCGCUAAGUUUC .....((((.(((((....))))).))))......((((..(((((((((....)))))))))............((((.....))))..(((.....)))......))))......... ( -42.20) >consensus UUUCAAGUUCGAACCAUGCGGUUCAAAAAAUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACGGGCAGGUUACCCACGUGUUACUCACCCGUCCGCCGCUAACUUCA .....((((.(((((....))))).))))......((((..((((.((((....)))).))))............((((.((....))..(((.....)))..))))))))......... (-31.72 = -29.32 + -2.40)

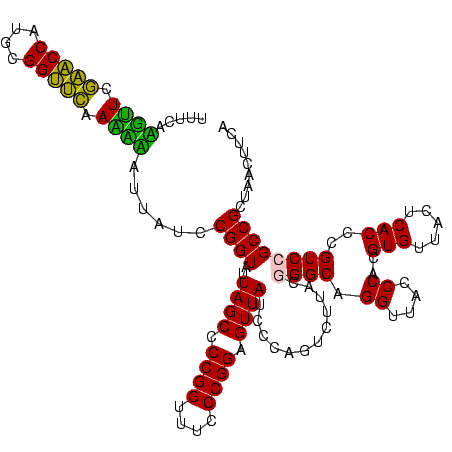
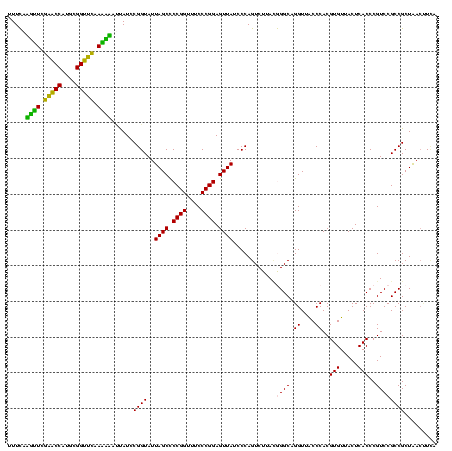
| Location | 30,194 – 30,313 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -54.48 |
| Consensus MFE | -56.58 |
| Energy contribution | -50.90 |
| Covariance contribution | -5.68 |
| Combinations/Pair | 1.50 |
| Mean z-score | -4.77 |
| Structure conservation index | 1.04 |
| SVM decision value | 3.69 |
| SVM RNA-class probability | 0.999531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 119 + 4214630/280-400 UGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUCAAACAUAAAAGGUGGCUUC-GGCUACCACUUACAGAUGGACCCGCGGCGC .(((((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((...-.)))))).))))))(.((....)))))).. ( -55.20) >Blich.0 34457 120 + 4222334/280-400 UGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUACAGAUGGACCCGCGGCGC .(((((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((.....)))))).))))))(.((....)))))).. ( -52.50) >Bcere.0 29012 119 + 5224283/280-400 UGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGC ...(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((...-.)))))).)))))))...(((....))).. ( -51.30) >Banth.0 29033 119 + 5227293/280-400 UGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGC ...(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((...-.)))))).)))))))...(((....))).. ( -51.20) >Bclau.0 52263 119 + 4303871/280-400 UGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAAUACCGGAUAAUCCCUUUCUCCACCUGGAGAGAGGGUGAAAGAUGGCUUC-GGCUAUCACUAGGGGAUGGGCCCGCGGCGC ...(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((...-.)))))).)))))))....(((...))).. ( -62.20) >consensus UGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAAUAGUUUGAACCGCAUGGUUCGAAAAUGAAAGGUGGCUUC_GGCUAUCACUUACGGAUGGACCCGCGGCGC .(((((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((.....)))))).)))))))..(....)))..... (-56.58 = -50.90 + -5.68)

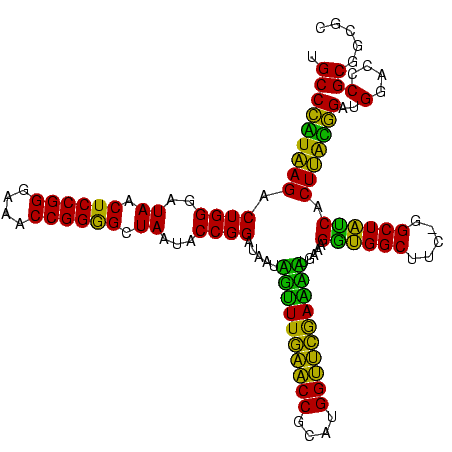
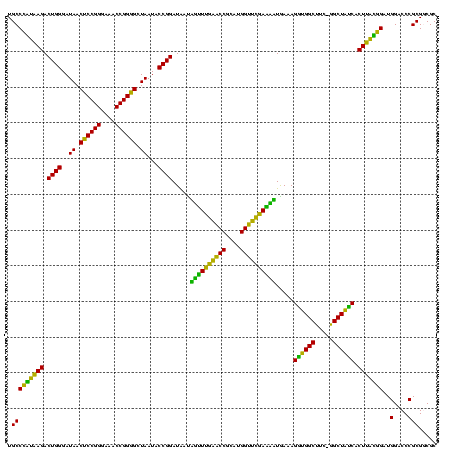
| Location | 30,194 – 30,313 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.67 |
| Mean single sequence MFE | -39.52 |
| Consensus MFE | -37.12 |
| Energy contribution | -32.84 |
| Covariance contribution | -4.28 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.599810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 119 + 4214630/280-400 GCGCCGCGGGUCCAUCUGUAAGUGGUAGCC-GAAGCCACCUUUUAUGUUUGAACCAUGCGGUUCAAACAACCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCA ..(((..((((..........(((((....-...)))))......((((((((((....))))))))))...))))((...((((.((((....)))).))))..)).........))). ( -40.20) >Blich.0 34457 120 + 4222334/280-400 GCGCCGCGGGUCCAUCUGUAAGUGGUAGCUAAAAGCCACCUUUUAUGAUUGAACCAUGCGGUUCAAUCAAGCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCA ..(((..((((.(....(((((.(((.((.....)).))).)))))(((((((((....)))))))))..).))))((...((((.((((....)))).))))..)).........))). ( -42.20) >Bcere.0 29012 119 + 5224283/280-400 GCGACGCGGGUCCAUCCAUAAGUGACAGCC-GAAGCCGCCUUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCA (((((....)))...((((((((((((...-..((((((..(((......)))....)))))).....)))))...((...((((.((((....)))).))))..))...))))))))). ( -35.70) >Banth.0 29033 119 + 5227293/280-400 GCGACGCGGGUCCAUCCAUAAGUGACAGCC-GAAGCCGCCUUUCAAUUUCGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCA (((((....)))...((((((((((.((.(-(....)).)).)))((((.(((((....))))).)))).......((...((((.((((....)))).))))..))...))))))))). ( -33.60) >Bclau.0 52263 119 + 4303871/280-400 GCGCCGCGGGCCCAUCCCCUAGUGAUAGCC-GAAGCCAUCUUUCACCCUCUCUCCAGGUGGAGAAAGGGAUUAUCCGGUAUUAGCUCCGGUUUCCCGGAGUUAUCCCAGUCUAAGGGGCA ((.((..((((..........(.(((((..-((((.....)))).((((.(((((....))))).)))).))))))((...(((((((((....)))))))))..)).))))..)).)). ( -45.90) >consensus GCGCCGCGGGUCCAUCCAUAAGUGAUAGCC_GAAGCCACCUUUCAAGUUCGAACCAUGCGGUUCAAAAAAUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACGGGCA .....(((....)..(((((((.(((.((.....)).))).....((((.(((((....))))).)))).......((...((((.((((....)))).))))..))...))))))))). (-37.12 = -32.84 + -4.28)


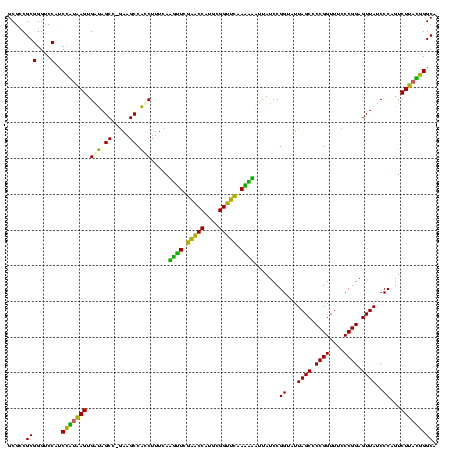
| Location | 30,194 – 30,313 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.34 |
| Mean single sequence MFE | -45.84 |
| Consensus MFE | -43.78 |
| Energy contribution | -39.18 |
| Covariance contribution | -4.60 |
| Combinations/Pair | 1.48 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.38 |
| SVM RNA-class probability | 0.999111 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 119 + 4214630/320-440 UACCGGAUGGUUGUUUGAACCGCAUGGUUCAAACAUAAAAGGUGGCUUC-GGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGA ..(((..((..((((((((((....)))))))))).....((((((...-.)))))).....))..)))...((((((......)))..((((((((.......)))))))).))).... ( -49.40) >Blich.0 34457 120 + 4222334/320-440 UACCGGAUGCUUGAUUGAACCGCAUGGUUCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGA ..(((..((..((((((((((....)))))))))).....((((((.....)))))).....))..)))...((((((......)))..((((((((.......)))))))).))).... ( -45.80) >Bcere.0 29012 119 + 5224283/320-440 UACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGA ..(((......((((((((((....)))))))))).....((((((...-.)))))).....)))((((((....))).)))..(((..((((((((.......)))))))))))..... ( -40.90) >Banth.0 29033 119 + 5227293/320-440 UACCGGAUAAUAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGA ..(((......((((((((((....)))))))))).....((((((...-.)))))).....)))((((((....))).)))..(((..((((((((.......)))))))))))..... ( -40.80) >Bclau.0 52263 119 + 4303871/320-440 UACCGGAUAAUCCCUUUCUCCACCUGGAGAGAGGGUGAAAGAUGGCUUC-GGCUAUCACUAGGGGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGA (((((......((((((((((....)))))))))).....((((((...-.))))))(((((..((((.(((...))).))))..)))))))))).(((((....)))))..(....).. ( -52.30) >consensus UACCGGAUAAUAGUUUGAACCGCAUGGUUCGAAAAUGAAAGGUGGCUUC_GGCUAUCACUUACGGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGA ..(((..((..((((((((((....)))))))))).....((((((.....)))))).....))..)))....((...((....))...((((((((.......)))))))).))..... (-43.78 = -39.18 + -4.60)

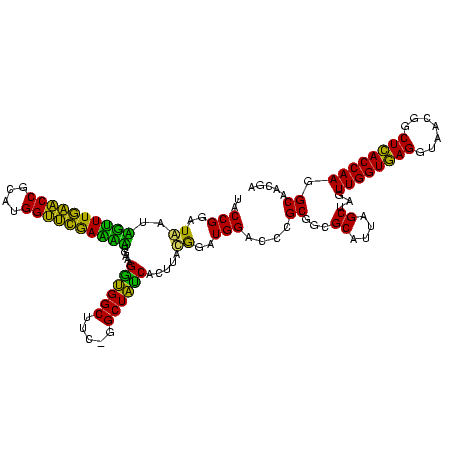

| Location | 30,194 – 30,313 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -52.18 |
| Consensus MFE | -45.76 |
| Energy contribution | -47.06 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.88 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998061 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 119 + 4214630/360-480 GGUGGCUUC-GGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((...-.)))))).............(((...(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))......))).. ( -53.50) >Blich.0 34457 120 + 4222334/360-480 GGUGGCUUUUAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((.....)))))).............(((...(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))......))).. ( -50.40) >Bcere.0 29012 119 + 5224283/360-480 GGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((...-.)))))).............(((((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).))...))).. ( -52.70) >Banth.0 29033 119 + 5227293/360-480 GGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((...-.)))))).............(((((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).))...))).. ( -52.70) >Bclau.0 52263 119 + 4303871/360-480 GAUGGCUUC-GGCUAUCACUAGGGGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((...-.)))))).((((.(....((((.((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....))))....)))).).))))... ( -51.60) >consensus GGUGGCUUC_GGCUAUCACUUACGGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGAC ((((((.....)))))).............(((.....(((((.(((..((((((((.......)))))))))))...)))))((.((((((((....)))...))))).))...))).. (-45.76 = -47.06 + 1.30)

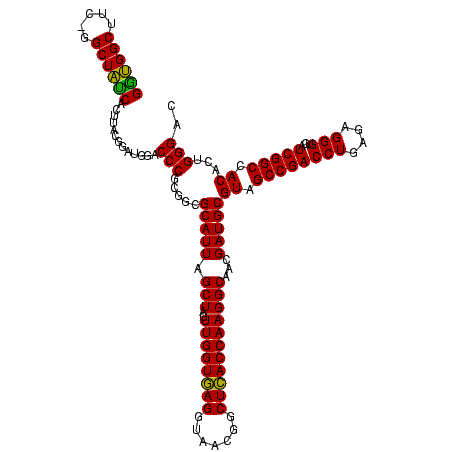
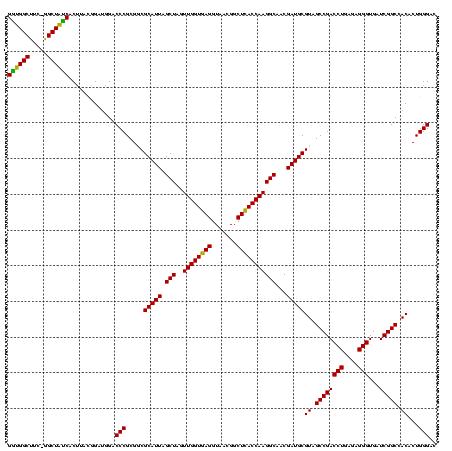
| Location | 30,194 – 30,313 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.30 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -40.42 |
| Energy contribution | -39.86 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 119 + 4214630/360-480 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCC-GAAGCCACC (.(((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).))))).).........(((((....-...))))). ( -46.80) >Blich.0 34457 120 + 4222334/360-480 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCUAAAAGCCACC (.(((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).))))).).........(((((........))))). ( -47.10) >Bcere.0 29012 119 + 5224283/360-480 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCC-GAAGCCGCC (.(((.((((((((((((.........)))))))))))).((((.((.((((((((.......))))))))...))....))))...))).).........(((...((.-...))))). ( -41.50) >Banth.0 29033 119 + 5227293/360-480 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCC-GAAGCCGCC (.(((.((((((((((((.........)))))))))))).((((.((.((((((((.......))))))))...))....))))...))).).........(((...((.-...))))). ( -41.50) >Bclau.0 52263 119 + 4303871/360-480 GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCGCCGCGGGCCCAUCCCCUAGUGAUAGCC-GAAGCCAUC ......((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))(((...((..(((....)))..-)).)))... ( -41.60) >consensus GUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCCAUAAGUGAUAGCC_GAAGCCACC (.(((.((((((((((((.........))))))))))))......((.((((((((.......))))))))...((....))...))))).)...........(((.((.....)).))) (-40.42 = -39.86 + -0.56)

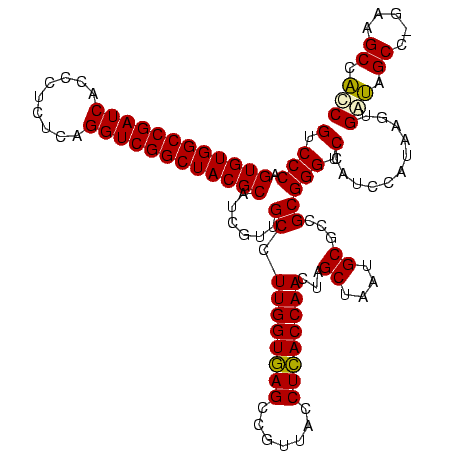

| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -51.18 |
| Consensus MFE | -46.50 |
| Energy contribution | -51.18 |
| Covariance contribution | 4.68 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.997800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/400-520 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.70) >Blich.0 34457 120 + 4222334/400-520 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.70) >Bcere.0 29012 120 + 5224283/400-520 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.60) >Banth.0 29033 120 + 5227293/400-520 AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -51.60) >Bclau.0 52263 120 + 4303871/400-520 AUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... ( -49.30) >consensus AUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGG ....(((..((((((((.......))))))))))).((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...)).... (-46.50 = -51.18 + 4.68)

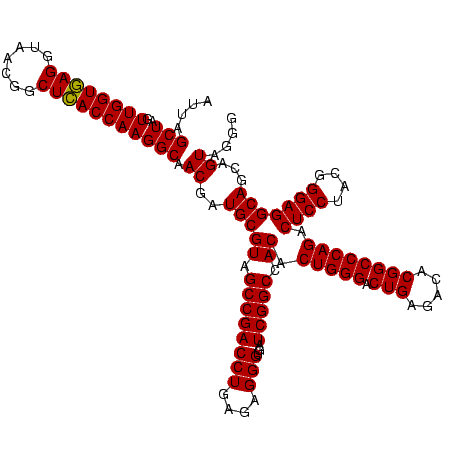
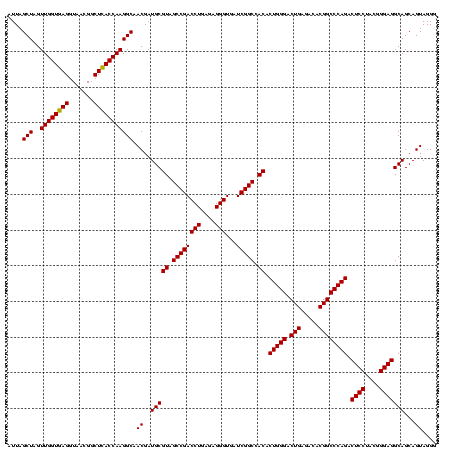
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.83 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -42.98 |
| Energy contribution | -42.66 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.84 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.992639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/400-520 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((((.......))))))))...)).... ( -43.30) >Blich.0 34457 120 + 4222334/400-520 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((((.......))))))))...)).... ( -43.30) >Bcere.0 29012 120 + 5224283/400-520 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))...((((..((((((((.......)))))))).)))).... ( -43.40) >Banth.0 29033 120 + 5227293/400-520 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))...((((..((((((((.......)))))))).)))).... ( -43.40) >Bclau.0 52263 120 + 4303871/400-520 CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))...((((..((((((((.......)))))))).)))).... ( -41.10) >consensus CCCUACUGCUGCCUCCCGUAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAU ....(((.((((.....)))).))).((((.(........).))))((((((((((((.........))))))))))))......((.((((((((.......))))))))...)).... (-42.98 = -42.66 + -0.32)

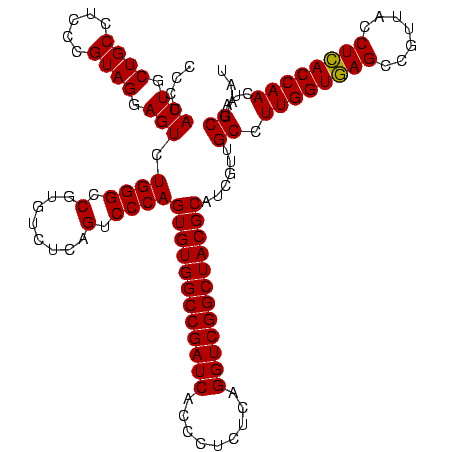
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -49.80 |
| Consensus MFE | -44.80 |
| Energy contribution | -49.80 |
| Covariance contribution | 5.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/440-560 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Blich.0 34457 120 + 4222334/440-560 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Bcere.0 29012 120 + 5224283/440-560 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Banth.0 29033 120 + 5227293/440-560 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >Bclau.0 52263 120 + 4303871/440-560 UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). ( -49.80) >consensus UGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCG (((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....))))...)))....((....(((((...(((((....))))).))))).....)). (-44.80 = -49.80 + 5.00)

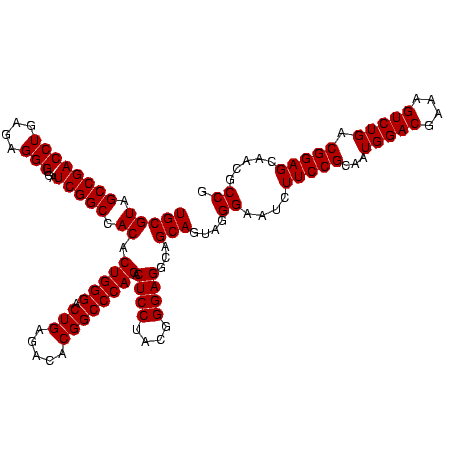
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.00 |
| Mean single sequence MFE | -48.80 |
| Consensus MFE | -44.30 |
| Energy contribution | -45.10 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.12 |
| Structure conservation index | 0.91 |
| SVM decision value | 5.00 |
| SVM RNA-class probability | 0.999968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/520-640 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUG ..((((((.....((((....))))((((((((..((.(((....))).))..((((....)))).....)))))))).....)))))).((((((((((((.....)))))))).)))) ( -48.90) >Blich.0 34457 120 + 4222334/520-640 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUG ..((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).((((((((((((.....)))))))).)))) ( -44.50) >Bcere.0 29012 120 + 5224283/520-640 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUG ..((((((.....((((....))))(((((((..(((((.((.((((.......)))).)))).)))....))))))).....)))))).((((((((((((.....)))))))).)))) ( -45.90) >Banth.0 29033 120 + 5227293/520-640 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUG ..((((((.....((((....))))(((((((..(((((.((.((((.......)))).)))).)))....))))))).....)))))).((((((((((((.....)))))))).)))) ( -45.90) >Bclau.0 52263 120 + 4303871/520-640 AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAGGAAGGCCUUCGGGUCGUAAAGCUCUGUUGUGAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUG ..((((((.((..((((....))))((((((((.....(((....))).....((((....)))).....)))))))).))..))))))...((((((((((.....))))))))))... ( -58.80) >consensus AAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUG ..((((((.....((((....))))(((((((...(..(((....)))..)..((((....))))......))))))).....))))))....(((((((((.....))))))))).... (-44.30 = -45.10 + 0.80)

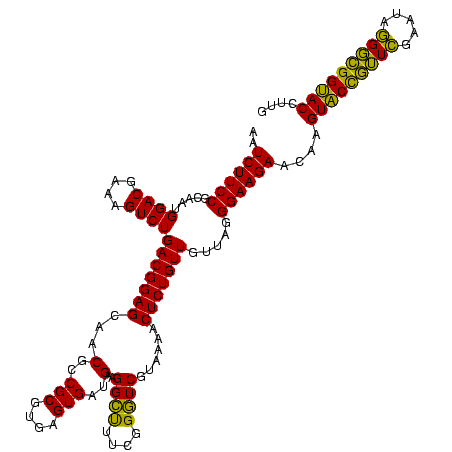
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -40.89 |
| Consensus MFE | -39.70 |
| Energy contribution | -37.58 |
| Covariance contribution | -2.12 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997876 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/560-680 CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG .(((.((......((((...(((((((....(((((....))))).....((((((((((((.....)))))))).))))))))).)).)))).....)))))(((.........))).. ( -37.90) >Blich.0 34457 120 + 4222334/560-680 CGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG .(((.((......((((...(((((((....(((((....))))).....((((((((((((.....)))))))).))))))))).)).)))).....)))))(((.........))).. ( -38.20) >Bcere.0 29012 120 + 5224283/560-680 CGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG (....).......(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..)))))))..(((.........))).. ( -40.90) >Banth.0 29033 120 + 5227293/560-680 CGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG (....).......(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..)))))))..(((.........))).. ( -40.90) >Bclau.0 52263 120 + 4303871/560-680 CGUGAGUGAGGAAGGCCUUCGGGUCGUAAAGCUCUGUUGUGAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUCACCAGAAAGCCACGGCUAACUACGUGCCAG .(((.((......((((....)))).......((((..((((((........((((((((((.....)))))))))).........))))))))))..)))))(((.........))).. ( -46.53) >consensus CGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAG .............(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..)))))))..(((.........))).. (-39.70 = -37.58 + -2.12)

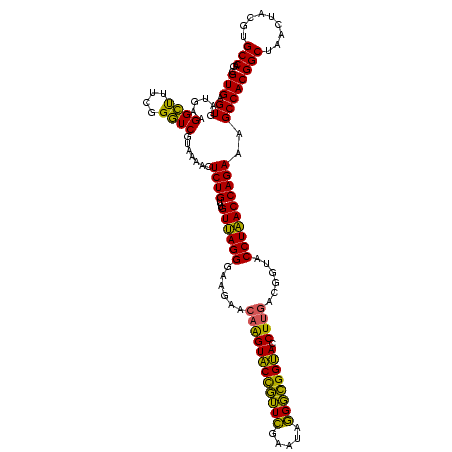
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -41.85 |
| Consensus MFE | -43.08 |
| Energy contribution | -40.84 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.83 |
| Structure conservation index | 1.03 |
| SVM decision value | 3.55 |
| SVM RNA-class probability | 0.999380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/600-720 UAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -41.40) >Blich.0 34457 120 + 4222334/600-720 UAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -41.40) >Bcere.0 29012 120 + 5224283/600-720 UAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUAUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -40.70) >Banth.0 29033 120 + 5227293/600-720 UAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUAUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -40.70) >Bclau.0 52263 120 + 4303871/600-720 GAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUCACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((........((((((((((.....)))))))))).........)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... ( -45.03) >consensus UAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGUUGUCCGGAAU ((((......((((((((((((.....)))))))).))))......)))).((.((.(((....((((.(((((((((.((....)))))...)))))).))))....))).)).))... (-43.08 = -40.84 + -2.24)

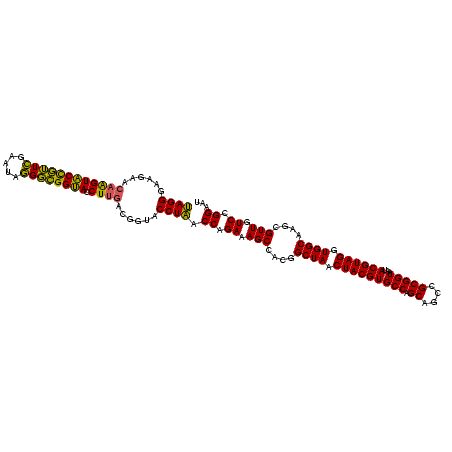
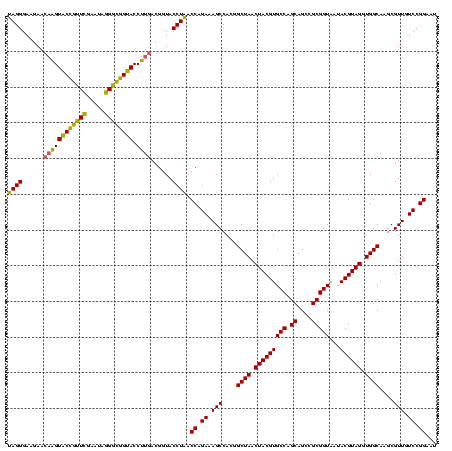
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -38.25 |
| Consensus MFE | -35.64 |
| Energy contribution | -35.96 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/600-720 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACUUGUUCUUCCCUA .....(((.((((..((((............((((((((((((.....))).)))))))))......(((..((((((.....)))))))))..........))))..))))..)))... ( -38.60) >Blich.0 34457 120 + 4222334/600-720 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACUUGUUCUUCCCUA .....(((.((((..((((............((((((((((((.....))).)))))))))......(((..((((((.....)))))))))..........))))..))))..)))... ( -38.60) >Bcere.0 29012 120 + 5224283/600-720 AUUCCGGAUAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUA .....(((.((((..(((.............((((((((((((.....))).)))))))))......((((.((((((.....))))))))))..........)))..))))..)))... ( -37.20) >Banth.0 29033 120 + 5227293/600-720 AUUCCGGAUAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUA .....(((.((((..(((.............((((((((((((.....))).)))))))))......((((.((((((.....))))))))))..........)))..))))..)))... ( -37.20) >Bclau.0 52263 120 + 4303871/600-720 AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUGAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACCGCUUCUUCCCUC .....(((....((....((((.........((((((((((((.....))).)))))))))......)))).(((((((((.(((......)))....).))))))))))....)))... ( -39.66) >consensus AUUCCGGACAACGCUUGCCACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACUUGUUCUUCCCUA .....(((.((((..(((((((.........((((((((((((.....))).)))))))))......)))..((((((.....)))))).............))))..))))..)))... (-35.64 = -35.96 + 0.32)

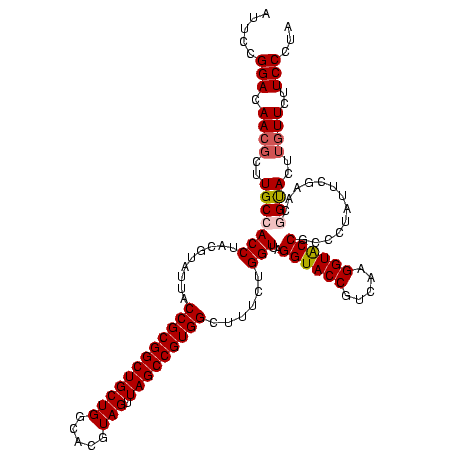
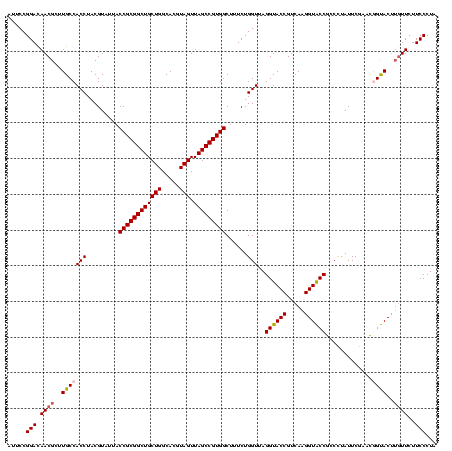
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.67 |
| Mean single sequence MFE | -39.38 |
| Consensus MFE | -38.88 |
| Energy contribution | -36.52 |
| Covariance contribution | -2.36 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/720-840 UAUUGGGCGUAAAGGGCUCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....((((.......))))(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))......((((.......)))). ( -37.80) >Blich.0 34457 120 + 4222334/720-840 UAUUGGGCGUAAAGCGCGCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....(.(((.....))).)(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))......((((.......)))). ( -39.20) >Bcere.0 29012 120 + 5224283/720-840 UAUUGGGCGUAAAGCGCGCGCAGGUGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCA ......(((.....)))((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))......((((.......)))). ( -38.10) >Banth.0 29033 120 + 5227293/720-840 UAUUGGGCGUAAAGCGCGCGCAGGUGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCA ......(((.....)))((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))......((((.......)))). ( -38.10) >Bclau.0 52263 120 + 4303871/720-840 UAUUGGGCGUAAAGCGCGCGCAGGCGGCUUCUUAAGUCUGAUGUGAAAUCUCGGGGCUCAACCCCGAGCGGCCAUUGGAAACUGGGGAGCUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....(.(((.....))).)(((..((((((((((..((..(((.(....(((((((.....)))))))...))))..))...)))))))).))..)))......((((.......)))). ( -43.70) >consensus UAUUGGGCGUAAAGCGCGCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCA ....(.(((.....))).)(((..((((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))).))..)))......((((.......)))). (-38.88 = -36.52 + -2.36)

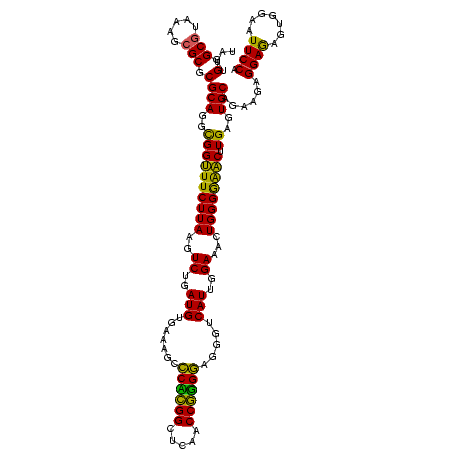
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -34.30 |
| Energy contribution | -32.18 |
| Covariance contribution | -2.12 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/760-880 AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA ........((((((((.....))))))((..((...(....)...))..)).....)).........(.(((...((((((((.(.(((......)))....).))))))))...)))). ( -35.80) >Blich.0 34457 120 + 4222334/760-880 AUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA ........((((((((.....))))))((..((...(....)...))..)).....)).........(.(((...((((((((.(.(((......)))....).))))))))...)))). ( -35.80) >Bcere.0 29012 120 + 5224283/760-880 AUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCA ........((((((((.....)))))).(((((...(....).....)))))....))...........(((...((((((((.(.(((......)))....).))))))))...))).. ( -32.20) >Banth.0 29033 120 + 5227293/760-880 AUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCA ........((((((((.....)))))).(((((...(....).....)))))....))...........(((...((((((((.(.(((......)))....).))))))))...))).. ( -32.20) >Bclau.0 52263 120 + 4303871/760-880 AUGUGAAAUCUCGGGGCUCAACCCCGAGCGGCCAUUGGAAACUGGGGAGCUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA ........((((((((.....))))((((..((...(....)..))..)))).............))))(((...((((((((.(.(((......)))....).))))))))...))).. ( -39.00) >consensus AUGUGAAAGCCCACGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCA .(((......((((((.....))))))((..((...(....)...))..)).....)))..........(((...((((((((.(.(((......)))....).))))))))...))).. (-34.30 = -32.18 + -2.12)

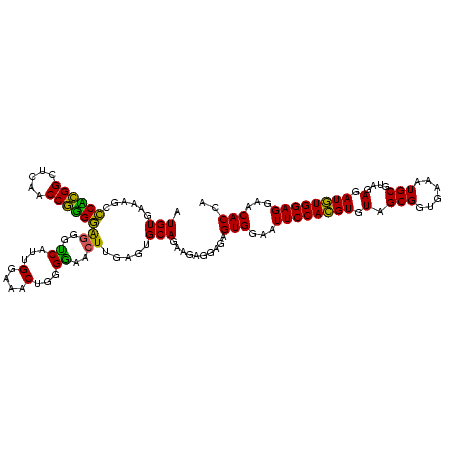
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.17 |
| Mean single sequence MFE | -24.31 |
| Consensus MFE | -24.12 |
| Energy contribution | -22.92 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.669454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/760-880 UGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAU .(((((................((........)).....((((....))))...........)))))...................(((...(((((((.....)))))))...)))... ( -25.40) >Blich.0 34457 120 + 4222334/760-880 UGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAU .(((((................((........)).....((((....))))...........)))))...................(((...(((((((.....)))))))...)))... ( -25.40) >Bcere.0 29012 120 + 5224283/760-880 UGGUGUUCCUCCAUAUCUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAU .((((.......................))))((.......((((.......))))......))......................(((...(((((((.....)))))))...)))... ( -21.52) >Banth.0 29033 120 + 5227293/760-880 UGGUGUUCCUCCAUAUCUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAU .((((.......................))))((.......((((.......))))......))......................(((...(((((((.....)))))))...)))... ( -21.52) >Bclau.0 52263 120 + 4303871/760-880 UGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGCUCCCCAGUUUCCAAUGGCCGCUCGGGGUUGAGCCCCGAGAUUUCACAU .((((.......................))))((((...((((....))))...........((......))..............))))..(((((((.....)))))))......... ( -27.70) >consensus UGGUGUUCCUCCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGGGGGCUUUCACAU .(((((................((........)).....((((....))))...........)))))...................(((...(((((((.....)))))))...)))... (-24.12 = -22.92 + -1.20)

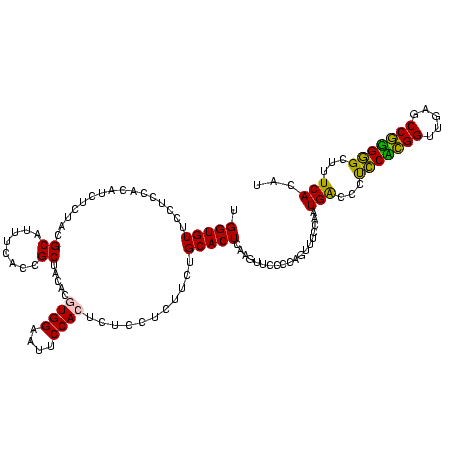
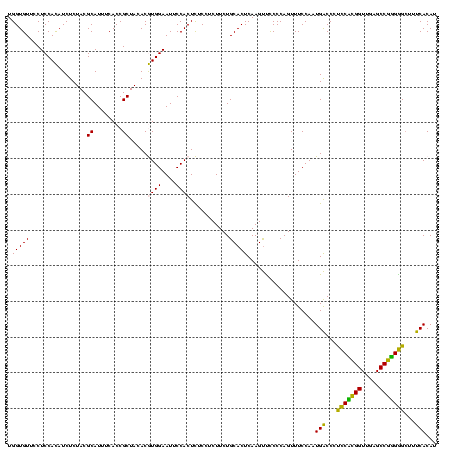
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.17 |
| Mean single sequence MFE | -39.14 |
| Consensus MFE | -39.22 |
| Energy contribution | -38.02 |
| Covariance contribution | -1.20 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.42 |
| Structure conservation index | 1.00 |
| SVM decision value | 4.69 |
| SVM RNA-class probability | 0.999939 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/800-920 ACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((...(.(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).).))).)). ( -40.00) >Blich.0 34457 120 + 4222334/800-920 ACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .((.((...(.(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).).))).)). ( -40.00) >Bcere.0 29012 120 + 5224283/800-920 ACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGG .(.((....)).)((((((((..(((((.(((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).......... ( -36.40) >Banth.0 29033 120 + 5227293/800-920 ACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGG .(.((....)).)((((((((..(((((.(((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).......... ( -36.40) >Bclau.0 52263 120 + 4303871/800-920 ACUGGGGAGCUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .......(((.(.((((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))).))).).))).... ( -42.90) >consensus ACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGG .........(((.(((((((.(((((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..)))....))).)))).))) (-39.22 = -38.02 + -1.20)

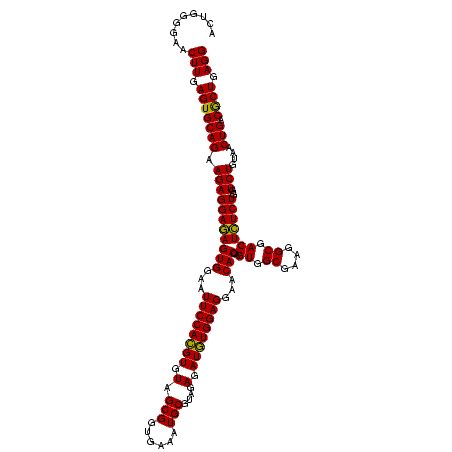
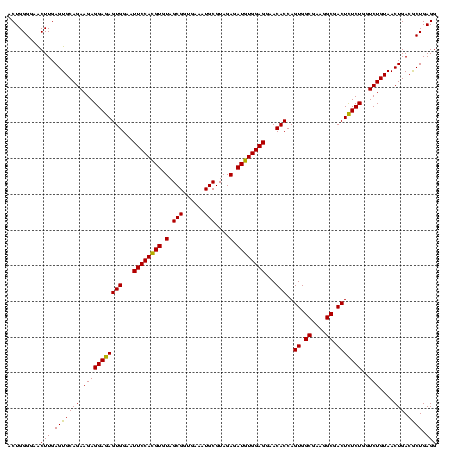
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.00 |
| Mean single sequence MFE | -40.44 |
| Consensus MFE | -40.20 |
| Energy contribution | -40.00 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/880-1000 GUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGAGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGU ((.((....)).))..............((((((((....((((...((((((...((...((((.........))))...))))))))((....)).....)))).))))))))..... ( -37.10) >Blich.0 34457 120 + 4222334/880-1000 GUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGU ...((....)).((((((((((.....)))((((((..(((((....((((((...((...((((.........))))...))))))))((....))....))))).))))))))))))) ( -41.20) >Bcere.0 29012 120 + 5224283/880-1000 GUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGU ...((....))((((....)))).....((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))..... ( -41.60) >Banth.0 29033 120 + 5227293/880-1000 GUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGU ...((....))((((....)))).....((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))..... ( -41.60) >Bclau.0 52263 120 + 4303871/880-1000 GUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUU ((.((....)).))..............((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))..... ( -40.70) >consensus GUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGU ...((....))((((....)))).....((((((((..(((((....(((((((..((...((((.........))))))..)))))))((....))....))))).))))))))..... (-40.20 = -40.00 + -0.20)

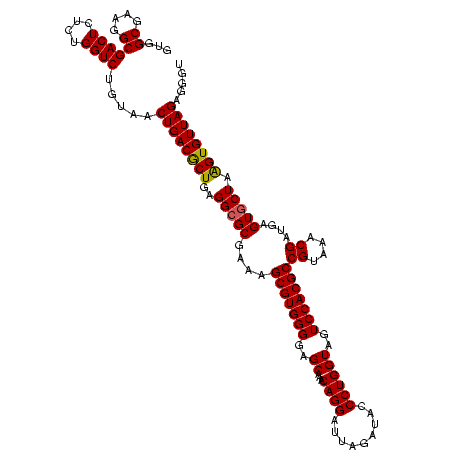
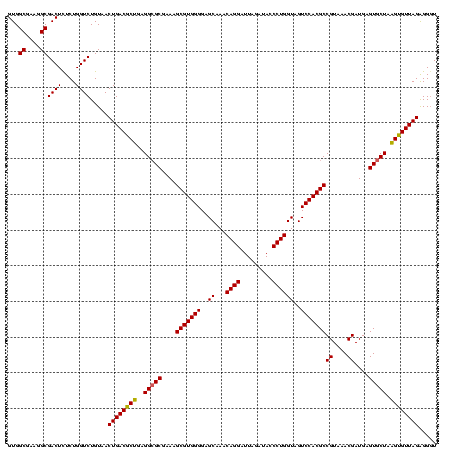
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -44.26 |
| Consensus MFE | -44.70 |
| Energy contribution | -43.78 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.35 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.94 |
| SVM RNA-class probability | 0.999963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/920-1040 AGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCC .(((...((((((...((...((((.........))))...))))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -48.90) >Blich.0 34457 120 + 4222334/920-1040 CGCGAAAGCGUGGGGAGCGAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCC .(((...((((((...((...((((.........))))...))))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -45.00) >Bcere.0 29012 120 + 5224283/920-1040 CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -42.40) >Banth.0 29033 120 + 5227293/920-1040 CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..(((((((((....)))))))))..))...((....))....))))))). ( -42.40) >Bclau.0 52263 120 + 4303871/920-1040 CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..(((.((((......)))).)))..))...((....))....))))))). ( -42.60) >consensus CGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCC .(((...(((((((..((...((((.........))))))..)))))))))).......(((((((.((..((((((((......))))))))..))...((....))....))))))). (-44.70 = -43.78 + -0.92)

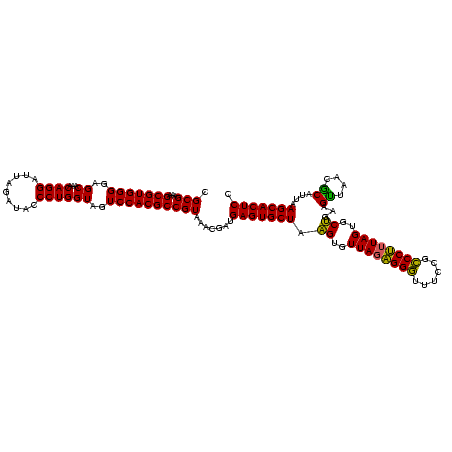

| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.50 |
| Mean single sequence MFE | -37.96 |
| Consensus MFE | -34.48 |
| Energy contribution | -33.72 |
| Covariance contribution | -0.76 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971567 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/920-1040 GGAGUGCUUAAUGCGUUAGCUGCAGCACUAAGGGGCGGAAACCCCCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGCUCCCCACGCUUUCGCU .(((((((....(.((((((....)).....((((.(....))))))))).)..))))))).........(((((((.....((((((.....))))))........)))))))...... ( -42.72) >Blich.0 34457 120 + 4222334/920-1040 GGAGUGCUUAAUGCGUUUGCUGCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUCGCUCCCCACGCUUUCGCG .(((((((....(.((((((....)))....((((.(....).))))))).)..)))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -37.12) >Bcere.0 29012 120 + 5224283/920-1040 GGAGUGCUUAAUGCGUUAACUUCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((((.(((..........))).)))((((.(....).))))........))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -35.72) >Banth.0 29033 120 + 5227293/920-1040 GGAGUGCUUAAUGCGUUAACUUCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((((.(((..........))).)))((((.(....).))))........))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -35.72) >Bclau.0 52263 120 + 4303871/920-1040 GGAGUGCUUAAUGUGUUAACUUCGGCACUACGGGCAUCGAAACCCCUAACACCUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((....((((((..((((((.(....)))..)))).....))))))..)))))))..(((....(((((((.....((((((.....))))))........)))))))...))) ( -38.52) >consensus GGAGUGCUUAAUGCGUUAACUUCAGCACUAAAGGGCGGAAACCCUCUAACACUUAGCACUCAUCGUUUACGGCGUGGACUACCAGGGUAUCUAAUCCUGUUUGCUCCCCACGCUUUCGCG .(((((((...(((..........)))....((((.(....).)))).......)))))))...((....(((((((.....((((((.....))))))........)))))))...)). (-34.48 = -33.72 + -0.76)

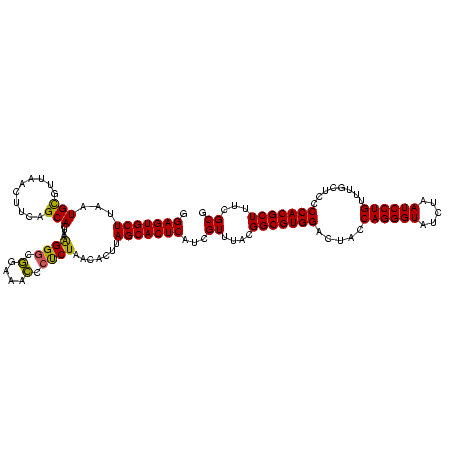
| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -47.70 |
| Consensus MFE | -48.74 |
| Energy contribution | -47.34 |
| Covariance contribution | -1.40 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.39 |
| Structure conservation index | 1.02 |
| SVM decision value | 6.32 |
| SVM RNA-class probability | 0.999998 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/960-1080 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -50.90) >Blich.0 34457 120 + 4222334/960-1080 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -47.00) >Bcere.0 29012 120 + 5224283/960-1080 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -46.80) >Banth.0 29033 120 + 5227293/960-1080 AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -46.80) >Bclau.0 52263 120 + 4303871/960-1080 AGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..(((.((((......)))).)))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ ( -47.00) >consensus AGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAU ..((((.((((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).)).))))((((.(((((....)))))..))))........ (-48.74 = -47.34 + -1.40)

| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -43.96 |
| Consensus MFE | -40.52 |
| Energy contribution | -40.40 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/1000-1120 UUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((((.((((((...((....))....))))))...(((...((((.(((((....)))))..))))..))).......))))))((((....)))).))))............. ( -48.80) >Blich.0 34457 120 + 4222334/1000-1120 UUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((...((((((...((....))....))))))((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).))))............. ( -44.20) >Bcere.0 29012 120 + 5224283/1000-1120 UUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((...((((((...((....))....))))))((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).))))............. ( -44.00) >Banth.0 29033 120 + 5227293/1000-1120 UUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ((((((((...((((((...((....))....))))))((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))).))))............. ( -44.00) >Bclau.0 52263 120 + 4303871/1000-1120 UCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCAGUGGAGCAUGUGGUUUAAU ......((((((((((..((((....)).))..)))))...(((...((((.(((((....)))))..))))..))).....)))))......((((.((......)).))))....... ( -38.80) >consensus UUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAU ....(((((..((((((...((....))....))))))...(((...((((.(((((....)))))..))))..)))........)))))...((((.((......)).))))....... (-40.52 = -40.40 + -0.12)

| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.00 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -40.20 |
| Energy contribution | -36.48 |
| Covariance contribution | -3.72 |
| Combinations/Pair | 1.34 |
| Mean z-score | -1.28 |
| Structure conservation index | 1.04 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826583 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/1400-1520 GACAGAACAAAGGGCAGCGAAACCGCGAGGUUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAG ((((((.....(((..((..((((....))))..))....))).....)))))).........((((((((((........))))))((((.(((((.....)))))..)))).)))).. ( -40.50) >Blich.0 34457 120 + 4222334/1400-1520 GGCAGAACAAAGGGCAGCGAAGCCGCGAGGCUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAG ((((((.....(((..((..((((....))))..))....))).....)))))).........((((((((((........))))))((((.(((((.....)))))..)))).)))).. ( -42.40) >Bcere.0 29012 120 + 5224283/1400-1520 GACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAG (.((((.......)))))..(((((((((((.(((.....))).....)))((((.(((((.....(((((((........)))))))...))))))))).........)))))).)).. ( -36.40) >Banth.0 29033 120 + 5227293/1400-1520 GACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAG (.((((.......)))))..(((((((((((.(((.....))).....)))((((.(((((.....(((((((........)))))))...))))))))).........)))))).)).. ( -36.40) >Bclau.0 52263 120 + 4303871/1400-1520 GAUGGUACAAAGGGCAGCGAAACCGCGAGGUGAAGCCAAUCCCAUAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAUUGCUAGUAAUCGCGGAUCAG ((((((.....(((..((...(((....)))...))....))).....)))))).....(((((..(((((((........))))))).....))))).((((.(....).))))..... ( -38.20) >consensus GACGGUACAAAGGGCAGCGAAACCGCGAGGUGAAGCCAAUCCCAUAAAACCGUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCUGGAAUCGCUAGUAAUCGCGGAUCAG ((((((.....(((..((...(((....)))...))....))).....))))))..(((((.....(((((((........)))))))...)))))((.((((........)))).)).. (-40.20 = -36.48 + -3.72)

| Location | 30,194 – 30,314 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.57 |
| Mean single sequence MFE | -42.38 |
| Consensus MFE | -39.88 |
| Energy contribution | -39.72 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 30194 120 + 4214630/1599-1719 GAGGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUUUACGG ((((((.((((..(((((((..(((((...))))).....((((((.......)))))).....))((((((((((....))))))))))......)))))....)))).)))))).... ( -43.20) >Blich.0 34457 119 + 4222334/1599-1719 GAGGUAACCUUUU-GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGG ..((((.((((((-(...((.(((......))).)).)))).((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))).))))...... ( -41.60) >Bcere.0 29012 119 + 5224283/1599-1719 GGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUGA ..(((..((....-)).)))..(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))............ ( -41.50) >Banth.0 29033 120 + 5227293/1599-1719 GGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUGA ..(((..((.....)).)))..(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))............ ( -41.50) >Bclau.0 52263 119 + 4303871/1599-1719 GAGGCAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGUUUUUACUC ..(((..((....-)).)))..(((((...)))))..((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))((((....)))) ( -44.10) >consensus GAGGUAACCUUUU_GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAUUUUACGA ..(((..((.....)).)))..(((((...))))).......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))................ (-39.88 = -39.72 + -0.16)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:20:31 2006