
| Sequence ID | Bclau.0 |
|---|---|
| Location | 2,861,663 – 2,861,783 |
| Length | 120 |
| Max. P | 0.999081 |

| Location | 2,861,663 – 2,861,776 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.74 |
| Mean single sequence MFE | -39.08 |
| Consensus MFE | -20.08 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.28 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.51 |
| SVM decision value | 3.36 |
| SVM RNA-class probability | 0.999081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 113 - 4303871/0-120 CCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGUUUUU-ACUCUAGUCGAUGUAU-----GACCUUCUGGUCAUAUAGCGCUU-GUGCUGUUGUUCAGUUUUGAGAGGACU (((((((....)))))))..........(((((((..((((((....-))))))(.((.(((((-----((((....))))))))).)))..-..((((.....))))...))))))).. ( -42.00) >Bhalo.0 4177891 120 - 4202352/0-120 CCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGGACCAUUCGCAGAUUCAUCUGCGGAGGGACGCUUCGGACUUUUGUUCAGUUUUGAAGGAAUU (((((((....)))))))..........((((((..(((((((.....)))))((.((..((.((((((((....))))))))))..))))..((((....)))).))...))))))... ( -44.00) >Banth.0 4655119 107 + 5227293/0-120 CCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UAAGCGCUGCUU------AUCAAUAUAAGUUUCCGUGUUUCG---UUUUGUUCAGUUUUGAGAGAACU (((((((....)))))))..(((.(((((((.......)))).(((((----(((((...))))------))))))..........))).)))(---((((.(((......)))))))). ( -35.20) >Bcere.0 4654098 107 + 5224283/0-120 CCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACGCUGUUC------AUCAAUAUAAGUUUCCGUGUUUCG---UUUUGUUCAGUUUUGAGAGAACU (((((((....)))))))..(((.(((((((.......)))).(((((----(((((...))))------))))))..........))).)))(---((((.(((......)))))))). ( -35.10) >consensus CCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA____UAAUCGCUGCAU______AUCAAUAUAAGUUUCCGCGCUUCG___UUUUGUUCAGUUUUGAGAGAACU (((((((....)))))))((((((...((((.......))))...........((((((.((.....................)).)))))).........))))))............. (-20.08 = -20.20 + 0.12)


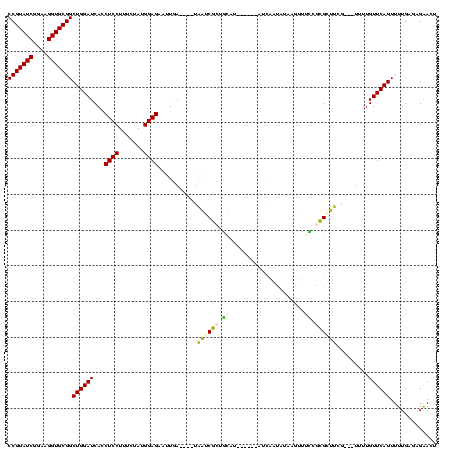
| Location | 2,861,663 – 2,861,777 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.29 |
| Mean single sequence MFE | -43.65 |
| Consensus MFE | -31.62 |
| Energy contribution | -31.62 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.75 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 114 - 4303871/40-160 AGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGUUUUU-ACUCUAGUCGAUGUAU-----GACCUUCUGGU ((((.(.(((..((((((((((((((...........((((((((((....))))))))))......((((.......)))).))))-)))))))))).))).)-----.))))...... ( -44.00) >Bhalo.0 4177891 120 - 4202352/40-160 AGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGGAGUUUUUAACUCUAGUCGGACCAUUCGCAGAUUCAUCUGC .(((..(((....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))...(((((.....))))).)))..)))....(((((....))))) ( -46.90) >Banth.0 4655119 110 + 5227293/40-160 AGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UAAGCGCUGCUU------AUCAAUAUAA .(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))(((.......)))..(((((----(((((...))))------)))))).... ( -41.90) >Bcere.0 4654098 110 + 5224283/40-160 AGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA----UGAACGCUGUUC------AUCAAUAUAA .(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))(((.......)))..(((((----(((((...))))------)))))).... ( -41.80) >consensus AGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGA____UAAUCGCUGCAU______AUCAAUAUAA .(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))(((.......)))....................................... (-31.62 = -31.62 + 0.00)

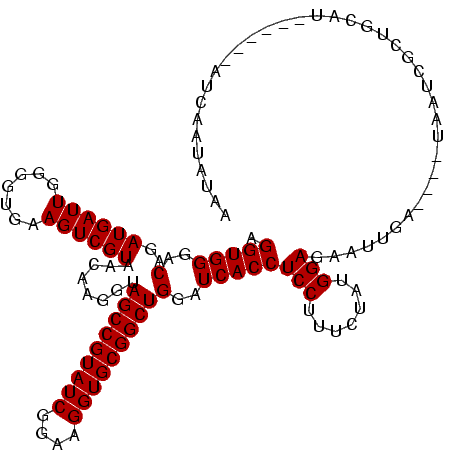
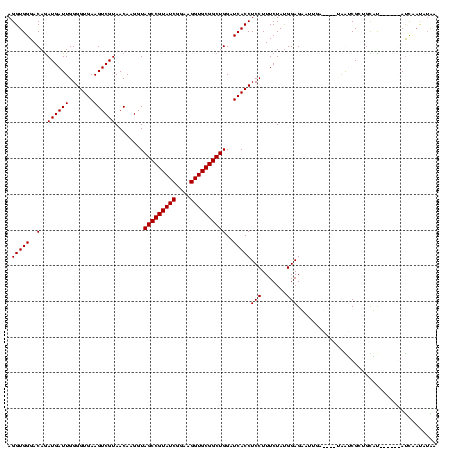
| Location | 2,861,663 – 2,861,782 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.91 |
| Mean single sequence MFE | -45.82 |
| Consensus MFE | -44.35 |
| Energy contribution | -43.98 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.614888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 119 - 4303871/80-200 CCGAAGUCGGUGAGGCAACC-UUUUGGAGCCAGCCGCCUAAGGUGGGACAAAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGG (((..(((.....(((..((-....)).)))..(((((...))))))))....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))) ( -46.10) >Bhalo.0 4177891 119 - 4202352/80-200 CCGAAGUCGGUGGGGUAACC-UUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUACGG (((..(((((((((....))-)......)))..(((((...))))))))....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))) ( -46.90) >Banth.0 4655119 120 + 5227293/80-200 CCGAAGUCGGUGGGGUAACCUUUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGG ((((((..(((......))).))))))..(((.(((((...))))).......((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))) ( -45.20) >Bcere.0 4654098 119 + 5224283/80-200 CCGAAGUCGGUGGGGUAACC-UUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGG ((..((.((((..(((..((-....)).))).)))).))..)).....((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..)). ( -45.10) >consensus CCGAAGUCGGUGGGGUAACC_UUUUGGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGG (((..(((.....(((..(......)..)))..(((((...))))))))....((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))) (-44.35 = -43.98 + -0.37)

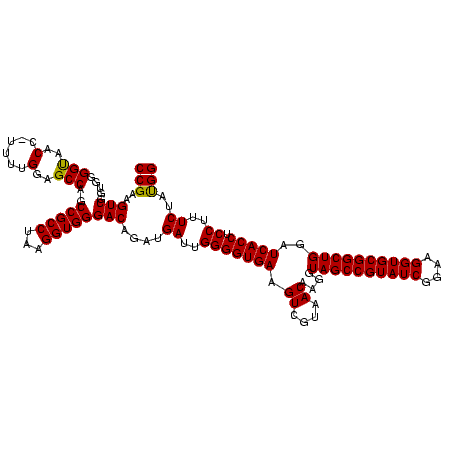
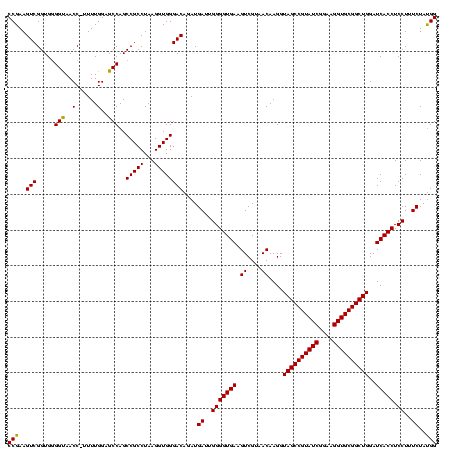
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -40.73 |
| Consensus MFE | -41.59 |
| Energy contribution | -39.90 |
| Covariance contribution | -1.69 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.87 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/280-400 CGUGCUACAAUGGAUGGUACAAAGGGCAGCGAAACCGCGAGGUGAAGCCAAUCCCAUAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAUUGCUAGUA ..((((((((((((((((.....(((..((...(((....)))...))....))).....)))))))....(((((..(((((((........))))))).....))))))))).))))) ( -44.50) >Bhalo.0 4177891 120 - 4202352/280-400 CGUGCUACAAUGGAUGGUACAAAGGGUUGCGAAGCCGCGAGGUGAAGCCAAUCCCAGAAAGCCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUGCAUGAAGCCGGAAUUGCUAGUA ..((((((((((((((((.....(((((((...(((....)))...).))))))......)))))))....(((((..(((((((........))))))).....))))))))).))))) ( -46.00) >Banth.0 4655119 120 + 5227293/280-400 CGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCUAGUA ..((((((...(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))....).))))) ( -36.20) >Bcere.0 4654098 120 + 5224283/280-400 CGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCGCUAGUA ..((((((...(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))....).))))) ( -36.20) >consensus CGUGCUACAAUGGACGGUACAAAGAGCUGCAAAACCGCGAGGUGAAGCCAAUCCCAUAAAACCAUUCUCAGUUCGGAUUGCAGGCUGCAACUCGCCUACAUGAAGCCGGAAUCGCUAGUA ..((((((((((((((((.....(((..((...(((....)))...))....))).....)))))))....(((((..(((((((........))))))).....))))))))).))))) (-41.59 = -39.90 + -1.69)

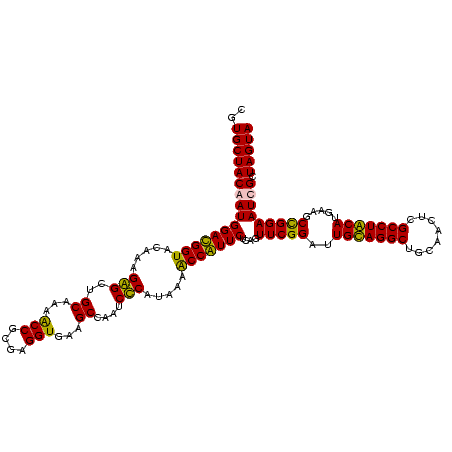
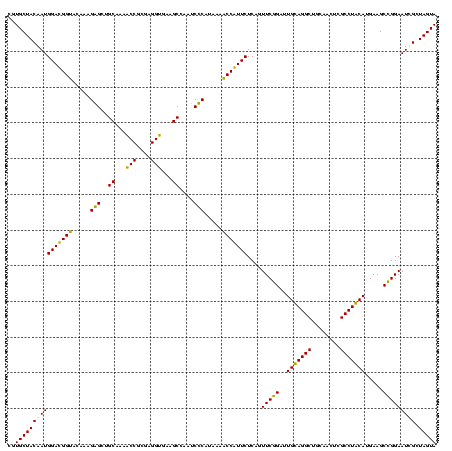
| Location | 2,861,663 – 2,861,781 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -39.05 |
| Consensus MFE | -37.35 |
| Energy contribution | -35.97 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.952148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 118 - 4303871/520-640 CGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUUUG-CCCCCGAAGGGGAA-GCCCAAUCUCUUGGGUGGUCAAAGGAUGUCAA .((((((...(((......)))....((....)).))))))((((((((.............)))).(((((-(((((....)))..-((((((....)))))))).)))))...)))). ( -39.42) >Bhalo.0 4177891 120 - 4202352/520-640 CGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUUUGUCCCCCGAAGGGGAAAGCCCUAUCUCUAGGGUGGUCAAAGGAUGUCAA .((((((...(((......)))....((....)).))))))((((((((.............)))).(((((.(((((....)))...((((((....)))))))).)))))...)))). ( -40.92) >Banth.0 4655119 118 + 5227293/520-640 CGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUCUG-CUCCCGAAGGAGAA-GCCCUAUCUCUAGGGUUGUCAGAGGAUGUCAA .((((((...(((......)))....((....)).))))))((((((((.............)))).(((((-((((....)))).(-((((((....)))))))..)))))...)))). ( -37.92) >Bcere.0 4654098 118 + 5224283/520-640 CGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUCUG-CUCCCGAAGGAGAA-GCCCUAUCUCUAGGGUUGUCAGAGGAUGUCAA .((((((...(((......)))....((....)).))))))((((((((.............)))).(((((-((((....)))).(-((((((....)))))))..)))))...)))). ( -37.92) >consensus CGCUCGUUGCGGGACUUAACCCAACAUCUCACGACACGAGCUGACGACAACCAUGCACCACCUGUCACUCUG_CCCCCGAAGGAGAA_GCCCUAUCUCUAGGGUGGUCAAAGGAUGUCAA .((((((...(((......)))....((....)).))))))((((((((.............)))).(((((.((((....))))...((((((....))))))...)))))...)))). (-37.35 = -35.97 + -1.38)

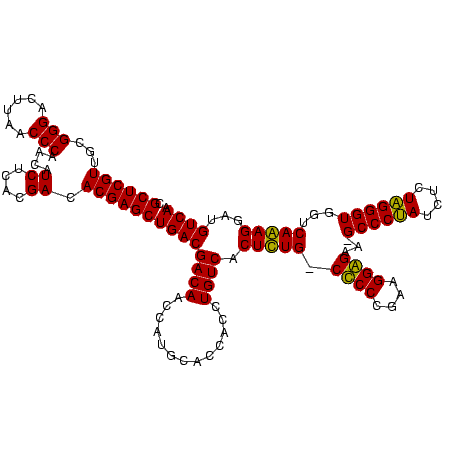
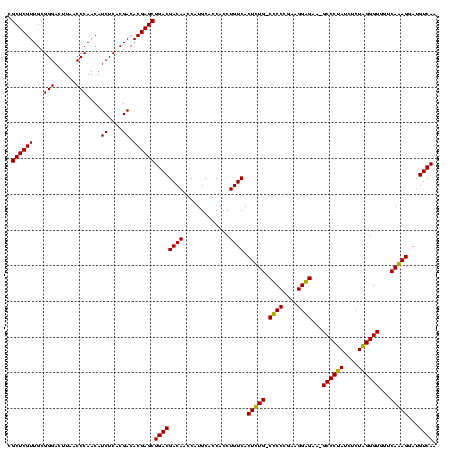
| Location | 2,861,663 – 2,861,781 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.52 |
| Mean single sequence MFE | -49.22 |
| Consensus MFE | -48.58 |
| Energy contribution | -47.20 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989800 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 118 - 4303871/520-640 UUGACAUCCUUUGACCACCCAAGAGAUUGGGC-UUCCCCUUCGGGGG-CAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCG ..((((.((..(((((((((((....))).((-((((((....))))-..))))....)))))).)).)).))))....(((((((((....)))((.(((......))))).)))))). ( -49.50) >Bhalo.0 4177891 120 - 4202352/520-640 UUGACAUCCUUUGACCACCCUAGAGAUAGGGCUUUCCCCUUCGGGGGACAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCG ..((((.((..(((((((((((....))).(((((((((....)))))..))))....)))))).)).)).))))....(((((((((....)))((.(((......))))).)))))). ( -51.80) >Banth.0 4655119 118 + 5227293/520-640 UUGACAUCCUCUGACAACCCUAGAGAUAGGGC-UUCUCCUUCGGGAG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCG .((((...(((((....(((((....))))).-..((((....))))-))))).(((((.((.....)).)))))))))(((((((((....)))((.(((......))))).)))))). ( -47.80) >Bcere.0 4654098 118 + 5224283/520-640 UUGACAUCCUCUGACAACCCUAGAGAUAGGGC-UUCUCCUUCGGGAG-CAGAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCG .((((...(((((....(((((....))))).-..((((....))))-))))).(((((.((.....)).)))))))))(((((((((....)))((.(((......))))).)))))). ( -47.80) >consensus UUGACAUCCUCUGACAACCCUAGAGAUAGGGC_UUCCCCUUCGGGAG_CAAAGUGACAGGUGGUGCAUGGUUGUCGUCAGCUCGUGUCGUGAGAUGUUGGGUUAAGUCCCGCAACGAGCG .((((...(((((....(((((....)))))....((((....)))).))))).(((((.((.....)).)))))))))(((((((((....)))((.(((......))))).)))))). (-48.58 = -47.20 + -1.38)

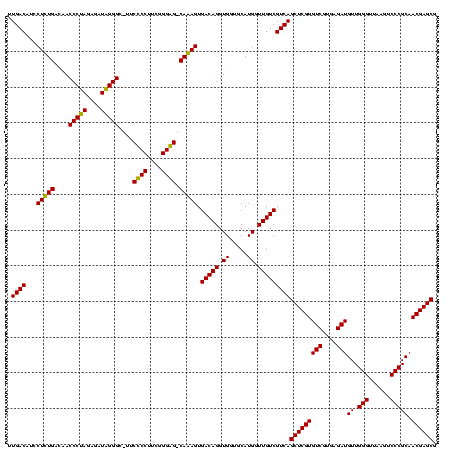
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -43.57 |
| Consensus MFE | -41.10 |
| Energy contribution | -40.23 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665385 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/680-800 GUUAGGGGUUUCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGACCCGCACAAGCAGUGGAGCAU (((..(((((......((((((((((..((((....)).))..)))))...(((...((((.(((((....)))))..))))..))).....)))))))))).(((.....))).))).. ( -40.40) >Bhalo.0 4177891 120 - 4202352/680-800 GUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAGUGGAGCAU ..(((((((......)))))))((((.................((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((.......)))).)))) ( -40.30) >Banth.0 4655119 120 + 5227293/680-800 GUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAU ..((((((((....))))))))(((((................((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))...))))) ( -46.80) >Bcere.0 4654098 120 + 5224283/680-800 GUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAU ..((((((((....))))))))(((((................((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))...))))) ( -46.80) >consensus GUUAGAGGGUUCCAGCCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCAGUGGAGCAU ..(((((((......)))))))((((.................((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((.......)))).)))) (-41.10 = -40.23 + -0.87)

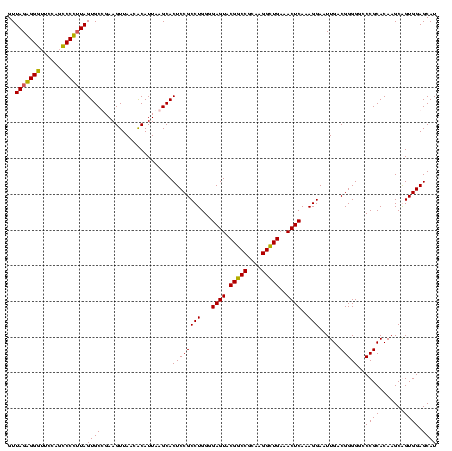
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -46.55 |
| Consensus MFE | -48.08 |
| Energy contribution | -46.20 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.54 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.94 |
| SVM RNA-class probability | 0.997815 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/720-840 AUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCGUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAAC ...(((.(((.........((....))..(((((((.((..(((.((((......)))).)))..))...((....))....))))))).))).))).....(((((....))))).... ( -46.90) >Bhalo.0 4177891 120 - 4202352/720-840 AUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGACCGCAAGGUUGAAAC ...(((.(((.........((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).))).))).....(((((....))))).... ( -45.90) >Banth.0 4655119 120 + 5227293/720-840 AUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAAC ...(((.(((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).))).....(((((....))))).... ( -46.70) >Bcere.0 4654098 120 + 5224283/720-840 AUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAAC ...(((.(((.........((....))..(((((((.((..(((((((((....)))))))))..))...((....))....))))))).))).))).....(((((....))))).... ( -46.70) >consensus AUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUCCAGCCCCUUAGUGCCGAAGUUAACACAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAAC ...(((.(((.........((....))..(((((((.((..((((((((......))))))))..))...((....))....))))))).))).))).....(((((....))))).... (-48.08 = -46.20 + -1.88)

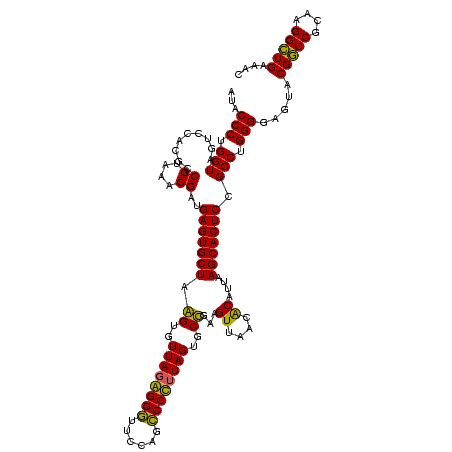
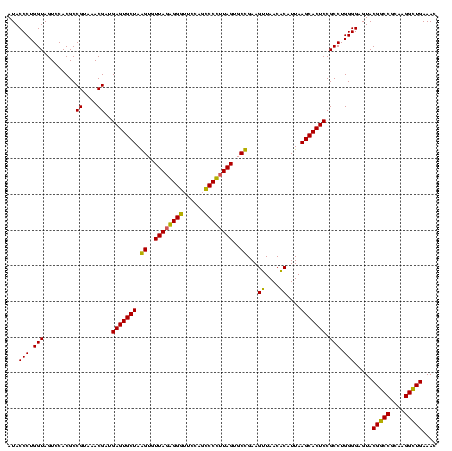
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -41.50 |
| Energy contribution | -40.25 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.36 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/760-880 GACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGAUGCCCGUAGUGCCGAAGUUAACACAU ..((....(((((....))(((((..((...((((.........))))))..)))))))).....))(((.(((((.((..(((.((((......)))).)))..))..)))..)).))) ( -40.30) >Bhalo.0 4177891 120 - 4202352/760-880 GACGCUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAGGUGUUAGGGGUUUCGACGCCCUUAGUGCCGAAGUUAACACAU ..((....(((((....))(((((..((...((((.........))))))..)))))))).....))(((.(((((.((..((((((((......))))))))..))..)))..)).))) ( -41.30) >Banth.0 4655119 120 + 5227293/760-880 GACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAU ........(((((....))(((((..((...((((.........))))))..)))))))).......(((.(((((.((..(((((((((....)))))))))..))..)))..)).))) ( -40.70) >Bcere.0 4654098 120 + 5224283/760-880 GACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAU ........(((((....))(((((..((...((((.........))))))..)))))))).......(((.(((((.((..(((((((((....)))))))))..))..)))..)).))) ( -40.70) >consensus GACACUGAGGCGCGAAAGCGUGGGGAGCAAACAGGAUUAGAUACCCUGGUAGUCCACGCCGUAAACGAUGAGUGCUAAGUGUUAGAGGGUUCCAGCCCCUUAGUGCCGAAGUUAACACAU ........(((((....))(((((..((...((((.........))))))..)))))))).......(((.(((((.((..((((((((......))))))))..))..)))..)).))) (-41.50 = -40.25 + -1.25)

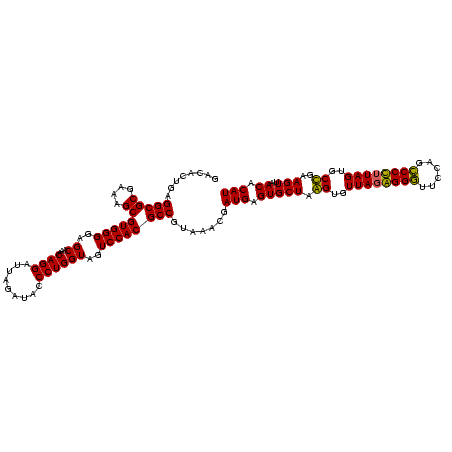
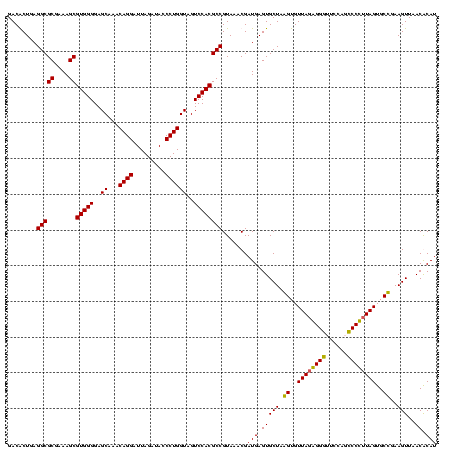
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.69 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -40.85 |
| Energy contribution | -39.10 |
| Covariance contribution | -1.75 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.93 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/880-1000 GCCAUUGGAAACUGGGGGGCUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACU (((...(....).....))).....((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))))... ( -43.70) >Bhalo.0 4177891 120 - 4202352/880-1000 GUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACU .(((..(....).((....))))).((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))))... ( -40.90) >Banth.0 4655119 120 + 5227293/880-1000 GUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACGCCAGUGGCGAAGGCGACUUUCUGGUCUGUAACU .(((..(....).((....))))).((((((...(((((((.(..((((((((.(.(((......)))....).))))))))...((((...))))....).)))))))..))))))... ( -37.20) >Bcere.0 4654098 120 + 5224283/880-1000 GUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACU .(((..(....).((....))))).((((((..(((((.(((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))))... ( -36.80) >consensus GUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACU (((...(....).....))).....((((((...((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).)))))))..))))))... (-40.85 = -39.10 + -1.75)

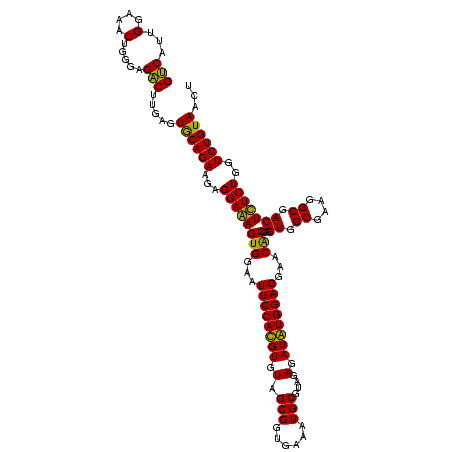
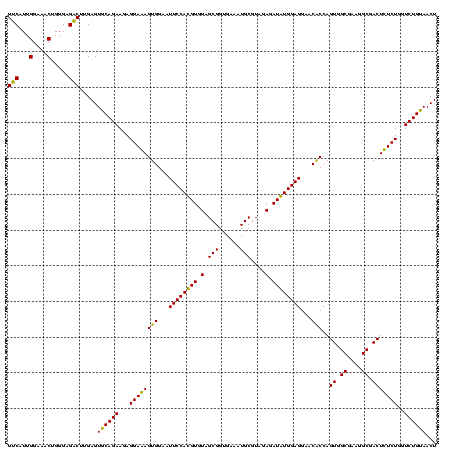
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.36 |
| Mean single sequence MFE | -24.35 |
| Consensus MFE | -22.33 |
| Energy contribution | -20.89 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697791 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/920-1040 CCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGCCCCCCAGUUUCCAAUGGCCGCUCGGGGUUGAGCCCCGAGAUUUCACAUCAGACUUAAG ............((........)).....((((....))))........((((.......(((..............)))..(((((((.....))))))).........))))...... ( -25.44) >Bhalo.0 4177891 120 - 4202352/920-1040 CCACAUCUCUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGUACUCAAGUCUCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAG .........(((((........)).....((((....))))...........)))((.((((((....(((......)))..(((((((.....)))))))..........)))))).)) ( -26.70) >Banth.0 4655119 120 + 5227293/920-1040 CCAUAUCUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAG ......................((.......((((.......))))......)).((.((((((....(((......)))..(((((((.....)))))))..........)))))).)) ( -22.62) >Bcere.0 4654098 120 + 5224283/920-1040 CCAUAUCUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAG ......................((.......((((.......))))......)).((.((((((....(((......)))..(((((((.....)))))))..........)))))).)) ( -22.62) >consensus CCACAUCUCUACGCAUUUCACCGCUACACAUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAG ............((........)).....((((....))))........((((.......(((..............)))..(((((((.....))))))).........))))...... (-22.33 = -20.89 + -1.44)

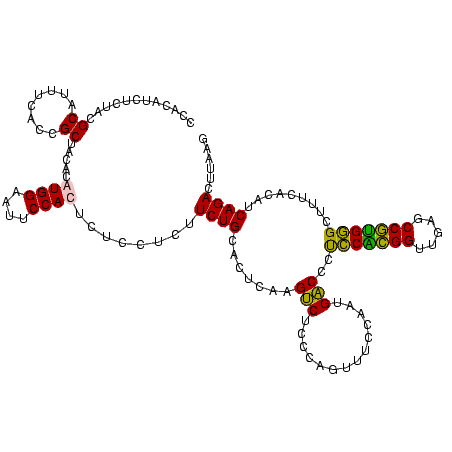
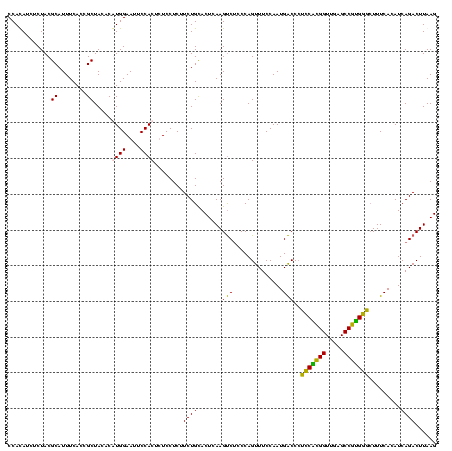
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.11 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -41.97 |
| Energy contribution | -39.48 |
| Covariance contribution | -2.50 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.50 |
| Structure conservation index | 1.03 |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.900679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/960-1080 UGUCCGGAAUUAUUGGGCGUAAAGCGCGCGCAGGCGGCUUCUUAAGUCUGAUGUGAAAUCUCGGGGCUCAACCCCGAGCGGCCAUUGGAAACUGGGGGGCUUGAGUGCAGAAGAGGAGAG (.(((.........(.(((.....))).)(((..((((((((((..((..(((.(....(((((((.....)))))))...))))..))...)))))))).))..)))......))).). ( -44.50) >Bhalo.0 4177891 120 - 4202352/960-1080 UGUCCGGAAUUAUUGGGCGUAAAGCGCGUGCAGGCGGUCUCUUAAGUCUGAUGUGAAAGCCCCCGGCUCAACCGGGGAGGGUCAUUGGAAACUGGGAGACUUGAGUACAGAAGAGGAGAG (.(((...........(((.....)))((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))......))).). ( -41.50) >Banth.0 4655119 120 + 5227293/960-1080 UAUCCGGAAUUAUUGGGCGUAAAGCGCGCGCAGGUGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAG ..(((...........(((.....)))((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))......)))... ( -38.80) >Bcere.0 4654098 120 + 5224283/960-1080 UAUCCGGAAUUAUUGGGCGUAAAGCGCGCGCAGGUGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAG ..(((...........(((.....)))((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))......)))... ( -38.80) >consensus UAUCCGGAAUUAUUGGGCGUAAAGCGCGCGCAGGCGGUUUCUUAAGUCUGAUGUGAAAGCCCACGGCUCAACCGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAG ..(((...........(((.....)))((((....(((((((((..((..(((.......((((((.....)))))).....)))..))...)))))))))...))))......)))... (-41.97 = -39.48 + -2.50)

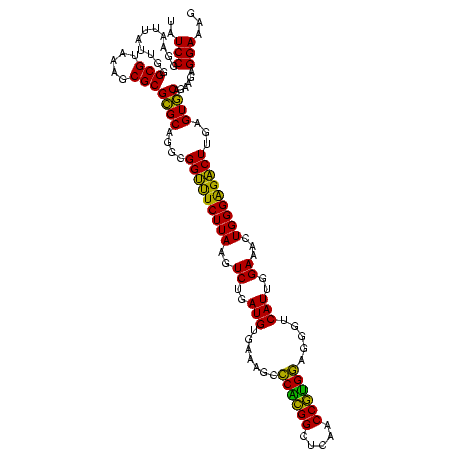
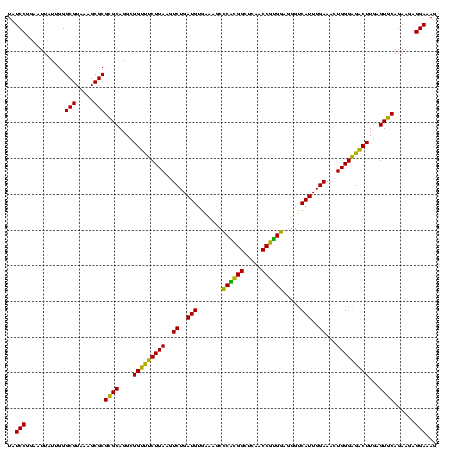
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.94 |
| Mean single sequence MFE | -46.68 |
| Consensus MFE | -45.74 |
| Energy contribution | -43.80 |
| Covariance contribution | -1.94 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998081 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1080-1200 GCUCUGUUGUGAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUCACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGU ((((((..((((((........((((((((((.....)))))))))).........))))))))))..(((((......(((((((((.((....)))))...)))))))))))..)).. ( -49.73) >Bhalo.0 4177891 119 - 4202352/1080-1200 ACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGU ....(.((((((((......((((((((((((...-.)))))))).))))......)))))))).)..((....((((.(((((((((.((....)))))...)))))).))))..)).. ( -49.00) >Banth.0 4655119 120 + 5227293/1080-1200 ACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGU ..((((..((((((......((((((((((((.....)))))))).))))......))))))))))..((....((((.(((((((((.((....)))))...)))))).))))..)).. ( -44.00) >Bcere.0 4654098 120 + 5224283/1080-1200 ACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGU ..((((..((((((......((((((((((((.....)))))))).))))......))))))))))..((....((((.(((((((((.((....)))))...)))))).))))..)).. ( -44.00) >consensus ACUCUGUUGUUAGGGAAGAACAAGUGCCAGUCGAAUAAGCCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUGGCAAGCGU ..((((..((((((......((((((((((((.....)))))))).))))......))))))))))..((....((((.(((((((((.((....)))))...)))))).))))..)).. (-45.74 = -43.80 + -1.94)

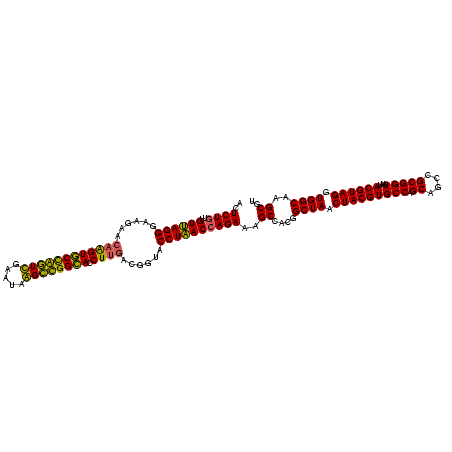
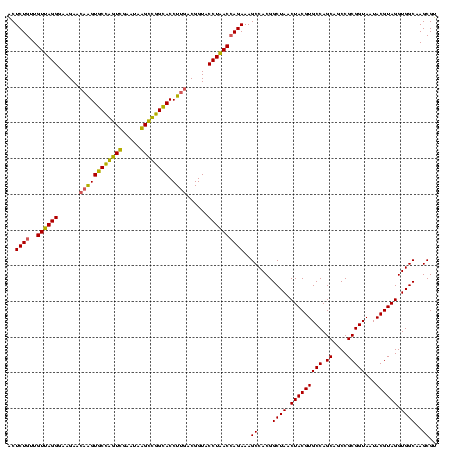
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -34.08 |
| Energy contribution | -33.09 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1120-1240 GUUAGCCGUGGCUUUCUGGUGAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACCGCUUCUUCCCUCACAACAGAGCUUUACGACCCGAAGGCCUUCCUCACUCACGCGGCGUUGCU ((..(((((((((((...((((((.........(((((((..(.....)..)))))))........))))))...)))))..........((.((....)))).....))))))...)). ( -39.83) >Bhalo.0 4177891 119 - 4202352/1120-1240 GUUAGCCGUGGCUUUCUCGUUAGGUACCGUCAAGGUGCCGCCCU-UUCGAACGGCACUUGUUCUUCCCUAACAACAGAGUUUUACGAUCCGAAAACCUUCAUCACUCACGCGGCGUUGCU ((..((((((........((((((........((((((((..(.-...)..)))))))).......))))))....((((.....(((..(((....)))))))))).))))))...)). ( -34.96) >Banth.0 4655119 120 + 5227293/1120-1240 GUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGACCCGAAAGCCUUCAUCACUCACGCGGCGUUGCU ((..((((((((((((.((((.((((((.....)))))).(((.........)))..............................)))).)))))))............)))))...)). ( -36.40) >Bcere.0 4654098 120 + 5224283/1120-1240 GUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGACCCGAAAGCCUUCAUCACUCACGCGGCGUUGCU ((..((((((((((((.((((.((((((.....)))))).(((.........)))..............................)))).)))))))............)))))...)). ( -36.40) >consensus GUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCACCCUAUUCAAACAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGACCCGAAAGCCUUCAUCACUCACGCGGCGUUGCU ((..((((((....((((((((((........(((((((((.(.....).))))))))).......))))))..))))(((((.((...)))))))............))))))...)). (-34.08 = -33.09 + -1.00)

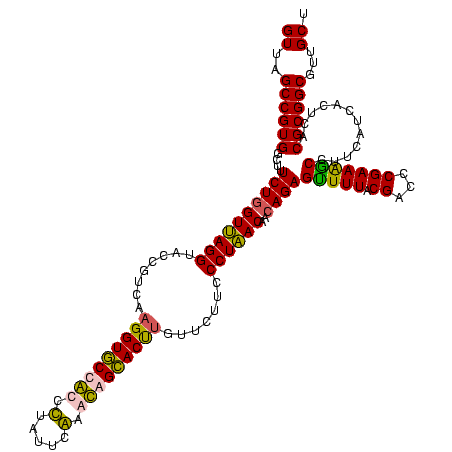
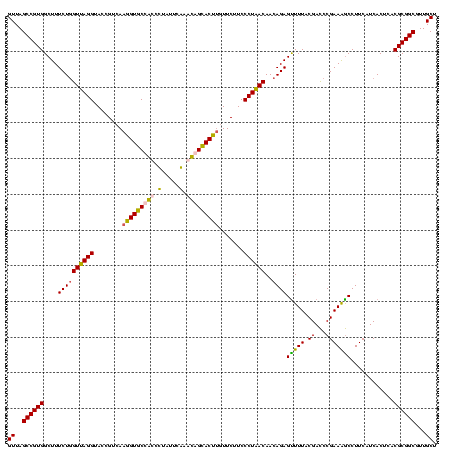
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -46.16 |
| Consensus MFE | -44.99 |
| Energy contribution | -42.92 |
| Covariance contribution | -2.06 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.16 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998907 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1120-1240 AGCAACGCCGCGUGAGUGAGGAAGGCCUUCGGGUCGUAAAGCUCUGUUGUGAGGGAAGAAGCGGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUCACCAGAAAGCCACGGCUAAC .((.....(((....))).....((((....)))).....))((((..((((((........((((((((((.....)))))))))).........)))))))))).(((....)))... ( -48.33) >Bhalo.0 4177891 119 - 4202352/1120-1240 AGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGGCGGCACCUUGACGGUACCUAACGAGAAAGCCACGGCUAAC ......(((((....))......((((((((((.........))).((((((((......((((((((((((...-.)))))))).))))......)))))))))))))))..))).... ( -47.10) >Banth.0 4655119 120 + 5227293/1120-1240 AGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAAC ......(((((....))......(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..)))))))..))).... ( -44.60) >Bcere.0 4654098 120 + 5224283/1120-1240 AGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAAC ......(((((....))......(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..)))))))..))).... ( -44.60) >consensus AGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCAGUCGAAUAAGCCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAAC ......(((((....))......(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..)))))))..))).... (-44.99 = -42.92 + -2.06)

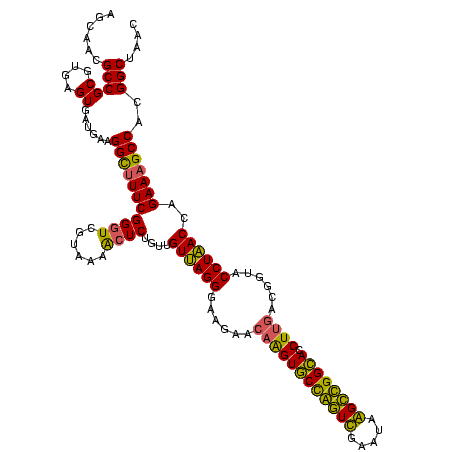
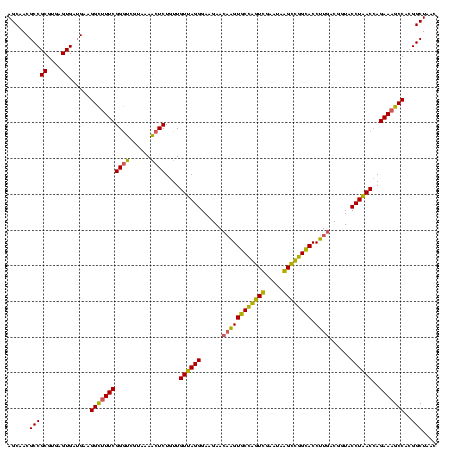
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.08 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -33.47 |
| Energy contribution | -33.23 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.695856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1160-1280 AGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAGGAAGGCCUUCGGGUCGUAAAGCUCUGUUGUGAGGGAAGAAGCGGUACCGUUCGAAUAGGG (((.(((..(..((((((.((..((((....))))((((((((.....(((....))).....((((....)))).....)))))))).))..))))))..)..))).)))......... ( -44.90) >Bhalo.0 4177891 119 - 4202352/1160-1280 AGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCGUUCGAA-AGGG .(((.(.(....((((((.....((((....))))(((((((...((.(((....))).))..((((....))))......))))))).....)))))).).).)))...((...-.)). ( -34.10) >Banth.0 4655119 120 + 5227293/1160-1280 AGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGC ((((.(.(....((((((.....((((....))))(((((((..(((((.((.((((.......)))).)))).)))....))))))).....)))))).).).))))............ ( -35.90) >Bcere.0 4654098 120 + 5224283/1160-1280 AGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGC ((((.(.(....((((((.....((((....))))(((((((..(((((.((.((((.......)))).)))).)))....))))))).....)))))).).).))))............ ( -35.90) >consensus AGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCCAGUCGAAUAAGC .(((.(.(....((((((.....((((....))))(((((((...(..(((....)))..)..((((....))))......))))))).....)))))).).).)))............. (-33.47 = -33.23 + -0.25)

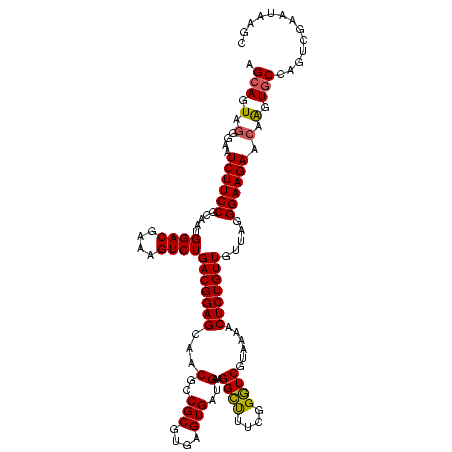
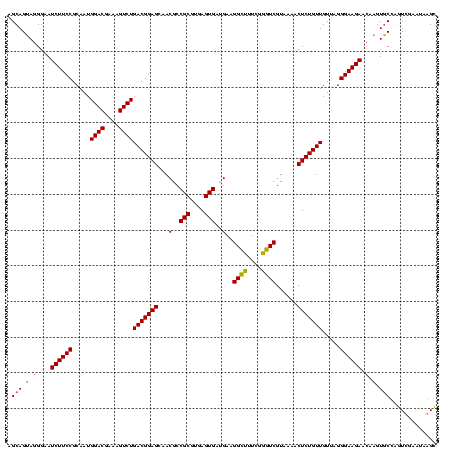
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 99.58 |
| Mean single sequence MFE | -48.72 |
| Consensus MFE | -48.72 |
| Energy contribution | -48.72 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.825970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1240-1360 AAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGG ...((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))...((((....))))..... ( -48.70) >Bhalo.0 4177891 120 - 4202352/1240-1360 AAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGG ...((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))...((((....))))..... ( -48.80) >Banth.0 4655119 120 + 5227293/1240-1360 AAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGG ...((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))...((((....))))..... ( -48.70) >Bcere.0 4654098 120 + 5224283/1240-1360 AAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGG ...((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))...((((....))))..... ( -48.70) >consensus AAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGCAGCAGUAGGGAAUCUUCCGCAAUGGACGAAAGUCUGACGG ...((.((..(((((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))))))...))..(((....)))))...((((....))))..... (-48.72 = -48.72 + 0.00)

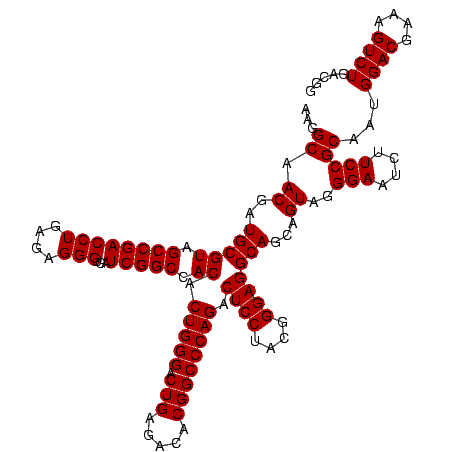
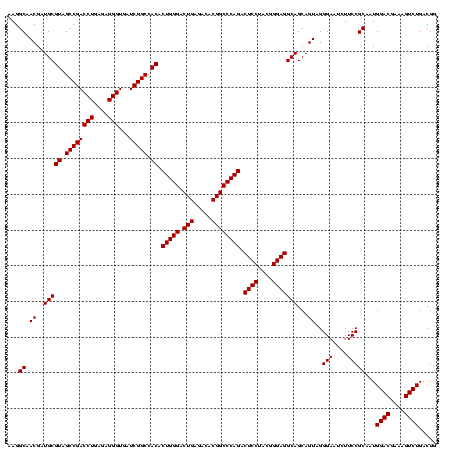
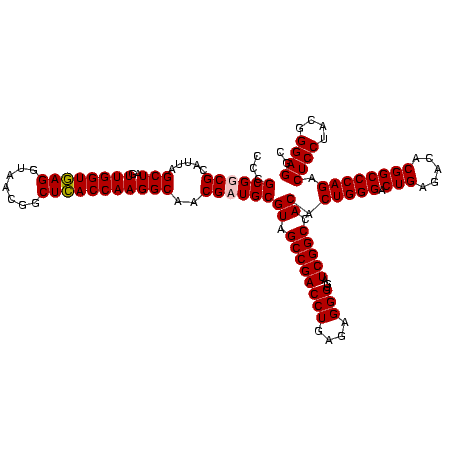
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.19 |
| Mean single sequence MFE | -53.90 |
| Consensus MFE | -52.58 |
| Energy contribution | -52.70 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929433 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1280-1400 CCCGCGGCGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGC ......(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))....(((((.(((.....)))))))).((((....)))).. ( -51.20) >Bhalo.0 4177891 120 - 4202352/1280-1400 CCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGC ......(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...)))))....(((((.(((.....)))))))).((((....)))).. ( -54.60) >Banth.0 4655119 120 + 5227293/1280-1400 CCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGC ...((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))).. ( -54.90) >Bcere.0 4654098 120 + 5224283/1280-1400 CCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGC ...((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))).. ( -54.90) >consensus CCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCCACACUGGGACUGAGACACGGCCCAGACUCCUACGGGAGGC ...((((((.....(((..((((((((.......)))))))))))..))))))((.((((((((....)))...))))).)).(((((.(((.....)))))))).((((....)))).. (-52.58 = -52.70 + 0.12)

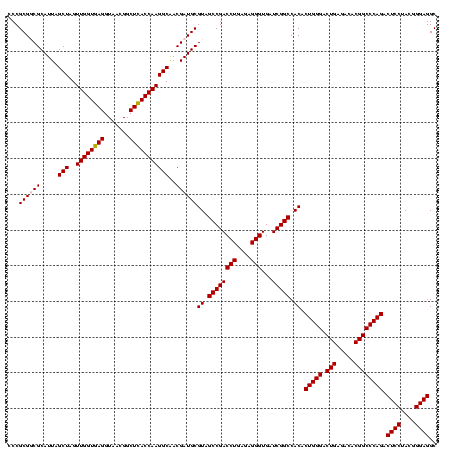
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -34.86 |
| Energy contribution | -34.55 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1320-1440 GGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUAAGCCGUUACCUUACCAACUAGCUAAUGCGCCGCGGGCCCAUCCCCUAGUGAUAGCCGAAGCCAUCUUUCACCCUCU (((.......((.....))((((((((((...((.((.((((((((.......))))))))...((....)))).))(((......)))..))).))))))).))).............. ( -36.20) >Bhalo.0 4177891 120 - 4202352/1320-1440 GGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGCCCAUCUGUAAGUGAUAGCCGAAACCAUCUUUCAAAAAAG (((..(((((.....(((((.((((.(((..(((.((.((((((((.......))))))))...))....)))..))))))).)))))...))))).)))((((.....))))....... ( -39.60) >Banth.0 4655119 120 + 5227293/1320-1440 GGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCGAAGCCGCCUUUCAAUUUCG (((.......((.....))((((((.(((.((((.((.((((((((.......))))))))...))....)))).))).((((........).))))))))).))).............. ( -37.90) >Bcere.0 4654098 120 + 5224283/1320-1440 GGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCGAAGCCGCCUUUCAAUUUCG (((.......((.....))((((((.(((.((((.((.((((((((.......))))))))...))....)))).))).((((........).))))))))).))).............. ( -37.90) >consensus GGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGCCCAUCCAUAAGUGACAGCCGAAGCCACCUUUCAAUUUCG ((((((((.........)))))))).(((.((((.((.((((((((.......))))))))...))....)))).)))(((.(((......)))...)))((((.....))))....... (-34.86 = -34.55 + -0.31)

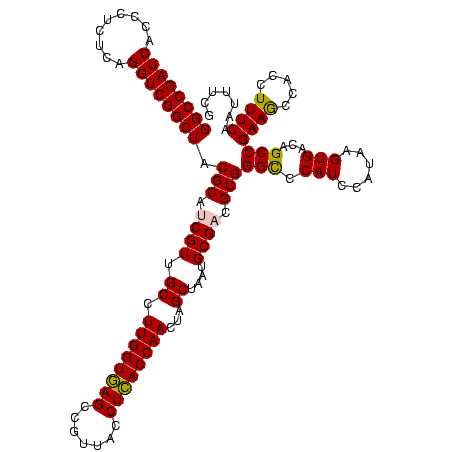
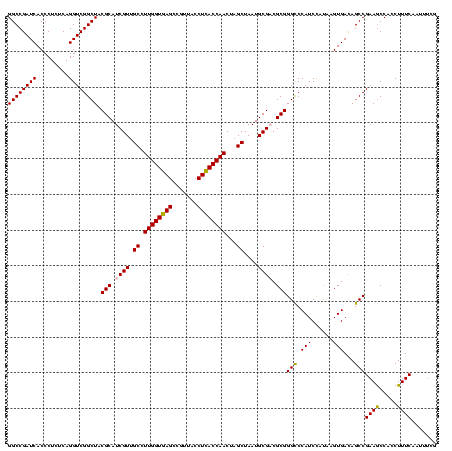
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -47.17 |
| Consensus MFE | -43.69 |
| Energy contribution | -42.88 |
| Covariance contribution | -0.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.907973 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1320-1440 AGAGGGUGAAAGAUGGCUUCGGCUAUCACUAGGGGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUAAGGUAACGGCUUACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCC .(.((((....((((((....)))))).(((.....))))))).).(((((((.(((..((((((((.......)))))))))))...))))))).((((((((....)))...))))). ( -50.50) >Bhalo.0 4177891 120 - 4202352/1320-1440 CUUUUUUGAAAGAUGGUUUCGGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCC ...........((((((....))))))...........((((.((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....))))....)))) ( -44.60) >Banth.0 4655119 120 + 5227293/1320-1440 CGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCC ...........((((((....))))))...............(((((((.....(((..((((((((.......)))))))))))..)))))))..((((((((....)))...))))). ( -46.80) >Bcere.0 4654098 120 + 5224283/1320-1440 CGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCC ...........((((((....))))))...............(((((((.....(((..((((((((.......)))))))))))..)))))))..((((((((....)))...))))). ( -46.80) >consensus CGAAAUUGAAAGACGGCUUCGGCUAUCACUUAUGGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGUGAUCGGCC ...........((((((....))))))...............(((((((.....(((..((((((((.......)))))))))))..)))))))..((((((((....)))...))))). (-43.69 = -42.88 + -0.81)


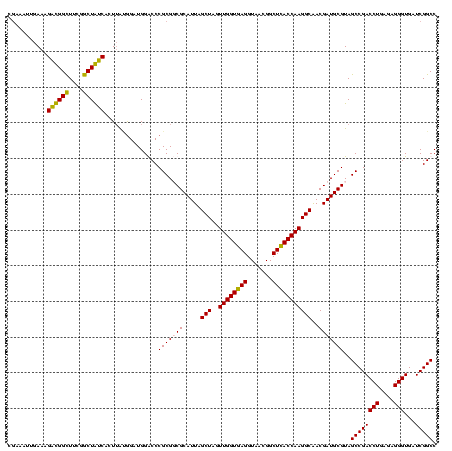
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -35.08 |
| Consensus MFE | -33.56 |
| Energy contribution | -29.88 |
| Covariance contribution | -3.69 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575148 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1360-1480 GGUAAGCCGUUACCUUACCAACUAGCUAAUGCGCCGCGGGCCCAUCCCCUAGUGAUAGCCGAAGCCAUCUUUCACCCUCUCUCCAGGUGGAGAAAGGGAUUAUCCGGUAUUAGCUCCGGU ((((((.......)))))).((((((((((..((((.((((...((..(((....)))..)).)))........((((.(((((....))))).)))).....))))))))))))..))) ( -39.70) >Bhalo.0 4177891 120 - 4202352/1360-1480 GGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGCCCAUCUGUAAGUGAUAGCCGAAACCAUCUUUCAAAAAAGAACCAUGCAGUUCUUUUUAUUAUCCGGUAUUAGCACCGAU ((((((.......)))))).....((((((..((((.((((..(((.......))).)))((((.....)))).(((((((((......))))))))).....)))))))))))...... ( -33.20) >Banth.0 4655119 120 + 5227293/1360-1480 GGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCGAAGCCGCCUUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGU ((((((.......)))))).....(((((((((((....))).........((((((.....((((((..(((......)))....)))))).....))))))...))))))))...... ( -33.70) >Bcere.0 4654098 120 + 5224283/1360-1480 GGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCCGAAGCCGCCUUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGU ((((((.......)))))).....(((((((((((....))).........((((((.....((((((..(((......)))....)))))).....))))))...))))))))...... ( -33.70) >consensus GGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGCCCAUCCAUAAGUGACAGCCGAAGCCACCUUUCAAUUUCGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCCCCGGU ((((((.......)))))).....((((((((......(((.(((......)))...)))((((.....)))).((((.(((((....))))).))))........))))))))...... (-33.56 = -29.88 + -3.69)

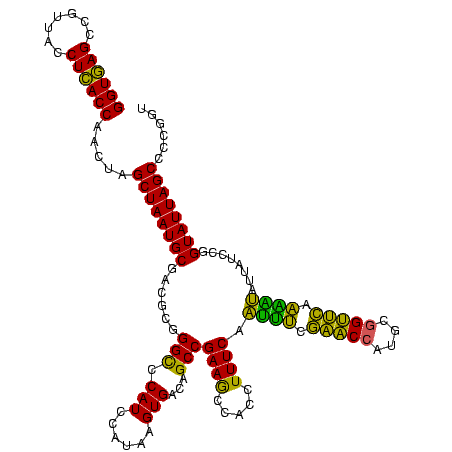
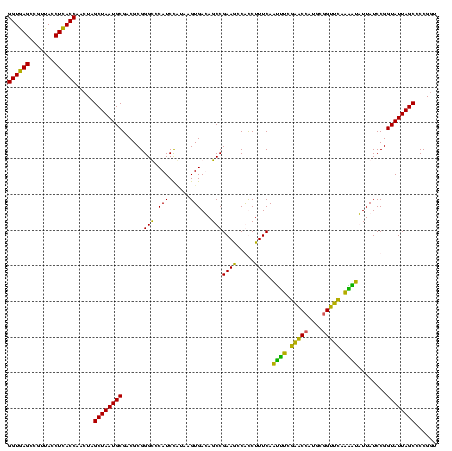
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -44.23 |
| Consensus MFE | -43.41 |
| Energy contribution | -39.23 |
| Covariance contribution | -4.19 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1360-1480 ACCGGAGCUAAUACCGGAUAAUCCCUUUCUCCACCUGGAGAGAGGGUGAAAGAUGGCUUCGGCUAUCACUAGGGGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUAAGGUAACGGCUUACC .((((........)))).....((((((((((....)))))))))).....((((((....))))))(((((..((((.(((...))).))))..))))).((((((.......)))))) ( -54.30) >Bhalo.0 4177891 120 - 4202352/1360-1480 AUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAAGAUGGUUUCGGCUAUCACUUACAGAUGGGCCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACC .((((((....)))))).....((((((((((....)))))))))).....((((((....))))))(((..(.((((.(((...))).)))).)..))).((((((.......)))))) ( -36.60) >Banth.0 4655119 120 + 5227293/1360-1480 ACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACC .((((........)))).....((((((((((....)))))))))).....((((((....))))))(((....(((((((....))).))))....))).((((((.......)))))) ( -43.00) >Bcere.0 4654098 120 + 5224283/1360-1480 ACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACC .((((........)))).....((((((((((....)))))))))).....((((((....))))))(((....(((((((....))).))))....))).((((((.......)))))) ( -43.00) >consensus ACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGACGGCUUCGGCUAUCACUUAUGGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACC .((((........)))).....((((((((((....)))))))))).....((((((....))))))(((....((((.(((...))).))))....))).((((((.......)))))) (-43.41 = -39.23 + -4.19)

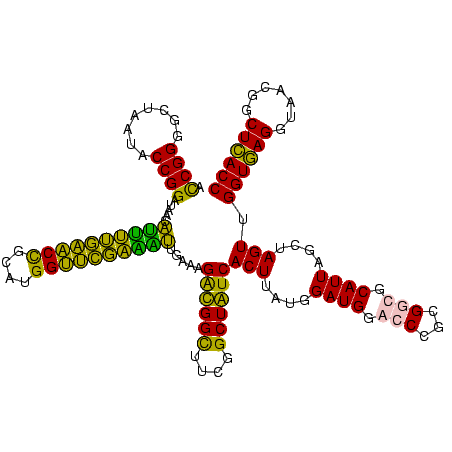
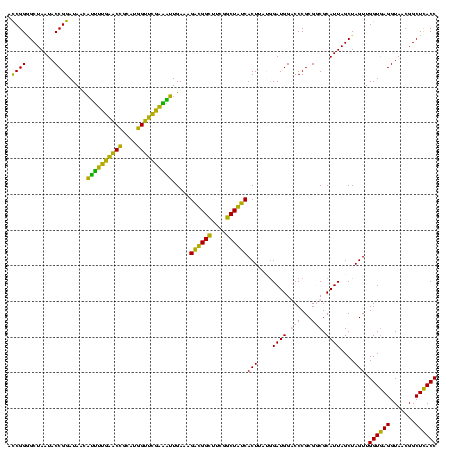
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -35.00 |
| Consensus MFE | -35.21 |
| Energy contribution | -30.52 |
| Covariance contribution | -4.69 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.56 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1400-1520 CCCAUCCCCUAGUGAUAGCCGAAGCCAUCUUUCACCCUCUCUCCAGGUGGAGAAAGGGAUUAUCCGGUAUUAGCUCCGGUUUCCCGGAGUUAUCCCAGUCUAAGGGGCAGGUUGCCCACG ...........(((.(((((...(((.((((.(.((((.(((((....))))).)))).......((...(((((((((....)))))))))..)).)...))))))).)))))..))). ( -45.60) >Bhalo.0 4177891 120 - 4202352/1400-1520 CCCAUCUGUAAGUGAUAGCCGAAACCAUCUUUCAAAAAAGAACCAUGCAGUUCUUUUUAUUAUCCGGUAUUAGCACCGAUUUCUCGAUGUUAUCCCAGUCUUACAGGCAGGUUGCCCACG ((..((((((((.(((((..((((.....)))).(((((((((......))))))))).))))).((...(((((.(((....))).)))))..))...))))))))..))......... ( -28.40) >Banth.0 4655119 120 + 5227293/1400-1520 UCCAUCCAUAAGUGACAGCCGAAGCCGCCUUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACG .((.(((((((((((((.....((((((..(((......)))....)))))).....)))))...((...((((.((((....)))).))))..))...))))))))..))......... ( -33.00) >Bcere.0 4654098 120 + 5224283/1400-1520 UCCAUCCAUAAGUGACAGCCGAAGCCGCCUUUCAAUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACG .((.(((((((((((((.....((((((..(((......)))....)))))).....)))))...((...((((.((((....)))).))))..))...))))))))..))......... ( -33.00) >consensus CCCAUCCAUAAGUGACAGCCGAAGCCACCUUUCAAUUUCGAACCAUGCGGUUCAAAAUAUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACG ((..((((((((((((....((((.....)))).((((.(((((....))))).))))))))...((...(((((((((....)))))))))..))...))))))))..))......... (-35.21 = -30.52 + -4.69)

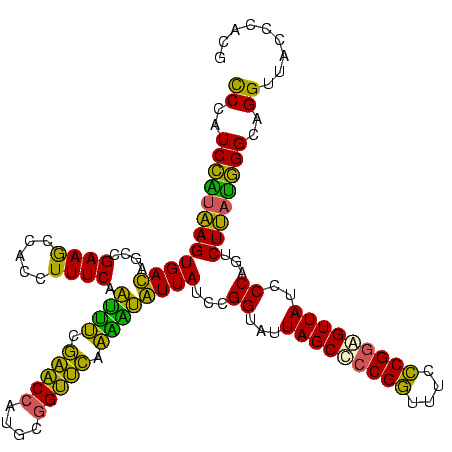
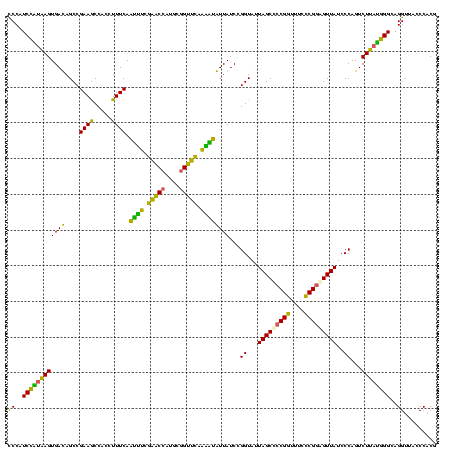
| Location | 2,861,663 – 2,861,783 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -52.60 |
| Consensus MFE | -59.30 |
| Energy contribution | -52.05 |
| Covariance contribution | -7.25 |
| Combinations/Pair | 1.61 |
| Mean z-score | -5.05 |
| Structure conservation index | 1.13 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 120 - 4303871/1400-1520 CGUGGGCAACCUGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAAUACCGGAUAAUCCCUUUCUCCACCUGGAGAGAGGGUGAAAGAUGGCUUCGGCUAUCACUAGGGGAUGGG .........(((..(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((....)))))).)))))))..))) ( -63.70) >Bhalo.0 4177891 120 - 4202352/1400-1520 CGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUUCUUUUUUGAAAGAUGGUUUCGGCUAUCACUUACAGAUGGG .........(((..(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((....)))))).)))))))..))) ( -41.10) >Banth.0 4655119 120 + 5227293/1400-1520 CGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGA .(..((...))..)(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((....)))))).)))))))..... ( -52.80) >Bcere.0 4654098 120 + 5224283/1400-1520 CGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGGCGGCUUCGGCUGUCACUUAUGGAUGGA .(..((...))..)(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((....)))))).)))))))..... ( -52.80) >consensus CGUGGGCAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAUUGAAAGACGGCUUCGGCUAUCACUUAUGGAUGGA .(..((...))..)(((((((.((((..((.((((((....)))))).))...)))).....((((((((((....)))))))))).....((((((....)))))).)))))))..... (-59.30 = -52.05 + -7.25)

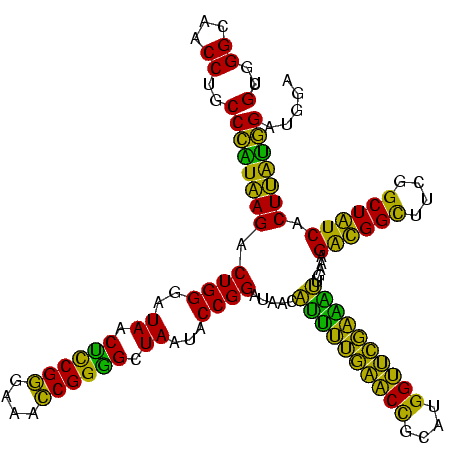
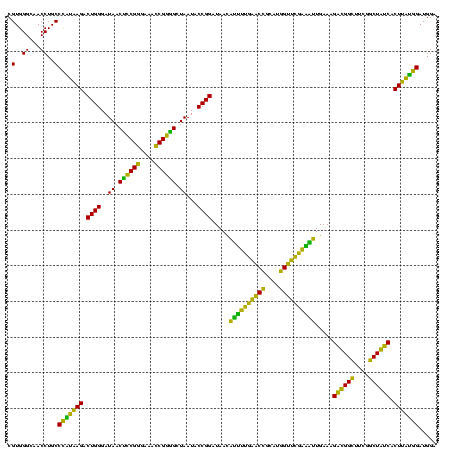
| Location | 2,861,663 – 2,861,782 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.95 |
| Mean single sequence MFE | -40.40 |
| Consensus MFE | -37.00 |
| Energy contribution | -35.00 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bclau.0 2861663 119 - 4303871/1440-1560 GCUUGCUCCCG-GACGUUAGCGGCGGACGGGUGAGUAACACGUGGGCAACCUGCCCCUUAGACUGGGAUAACUCCGGGAAACCGGAGCUAAUACCGGAUAAUCCCUUUCUCCACCUGGAG ((((...((((-..(((.....)))..)))).)))).....(.((((.....))))).....((((..((.((((((....)))))).))...))))...........((((....)))) ( -46.20) >Bhalo.0 4177891 119 - 4202352/1440-1560 GCUUGCUCCUA-GAGGUUAGCGGCGAACGGGUGAGUAACACGUGGGCAACCUGCCUGUAAGACUGGGAUAACAUCGAGAAAUCGGUGCUAAUACCGGAUAAUAAAAAGAACUGCAUGGUU ((((((((.((-.((((..(..((..(((.((.....)).)))..))..)..)))).)).))((((..((.((((((....)))))).))...))))...............))).))). ( -32.60) >Banth.0 4655119 120 + 5227293/1440-1560 GCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUU (((.((((....).))).))).((((..((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............))))...... ( -41.40) >Bcere.0 4654098 120 + 5224283/1440-1560 GCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUU (((.((((....).))).))).((((..((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............))))...... ( -41.40) >consensus GCUUGCUCCUA_GAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGCAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUU (((.((((....)).)).)))((((.(((.((.....)).)))((....)))))).......((((..((.((((((....)))))).))...))))...........((((....)))) (-37.00 = -35.00 + -2.00)

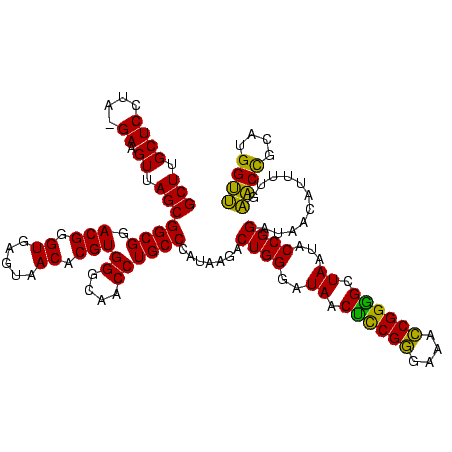
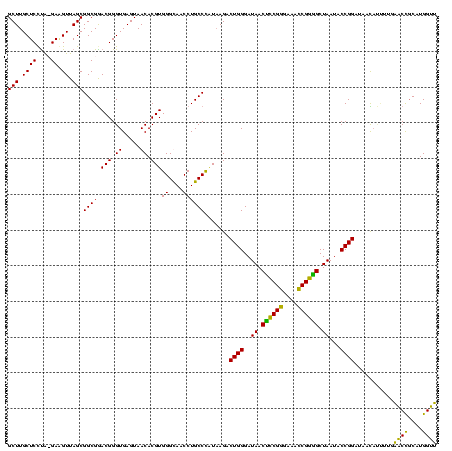
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:20:04 2006