
| Sequence ID | Banth.0 |
|---|---|
| Location | 4,651,543 – 4,651,652 |
| Length | 109 |
| Max. P | 0.998448 |

| Location | 4,651,543 – 4,651,652 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -34.05 |
| Energy contribution | -32.83 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.86 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4651543 109 + 5227293/0-120 GUUAUUUAA------UUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCCA---AAGGCGCCAGAUCCUAAGUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUAUUAA--UUUAU .......((------((((..(((((((((..(((.(.....).)))..(.((---.(.(((((((((.....))))))))).).)).)..........))))))))).))))--))... ( -38.90) >Bcere.0 4650521 109 + 5224283/0-120 GUUAUUUAA------UUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCCA---AAGGCGCCAGAUCCUAAGUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUAUUAA--UUUAU .......((------((((..(((((((((..(((.(.....).)))..(.((---.(.(((((((((.....))))))))).).)).)..........))))))))).))))--))... ( -38.90) >Bclau.0 2626639 120 + 4303871/0-120 GUUGCUUAAAGAGUUUUUGAAAUGCGGGUGAAGGGACUUGAACCCCCACGUCCGUUAGGACACUAGAGCCUGAUUCUAGCGCGUCUGCCAAUUCCGCCACACCCGCAUACCAAAGCUCAG ..........(((((((.(..(((((((((..((((.(((.........((((....))))..(((((((((....))).)).)))).))).))).)..)))))))))..)))))))).. ( -41.00) >consensus GUUAUUUAA______UUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCCA___AAGGCGCCAGAUCCUAAGUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUAUUAA__UUUAU .....................(((((((((..(((.........)))..(((......)))(((((((.....)))))))(((...........)))..)))))))))............ (-34.05 = -32.83 + -1.22)

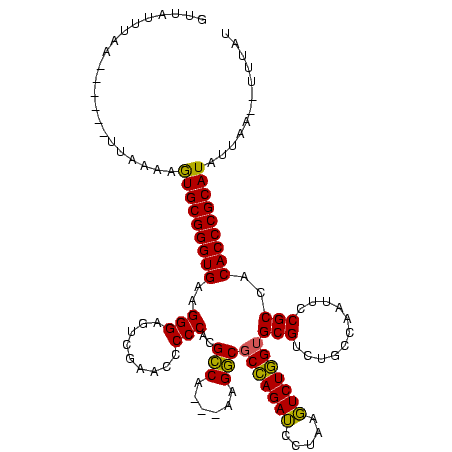
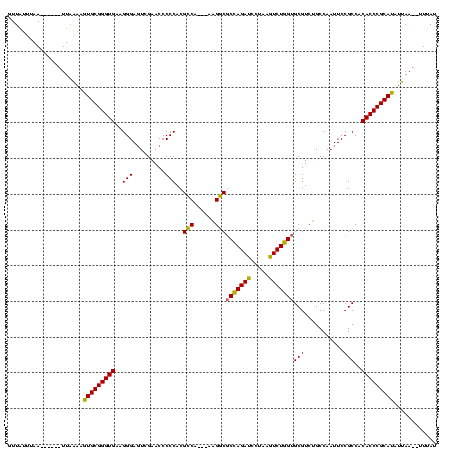
| Location | 4,651,543 – 4,651,652 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -38.74 |
| Energy contribution | -36.97 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.00 |
| SVM RNA-class probability | 0.998093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4651543 109 + 5227293/0-120 AUAAA--UUAAUAUGCGGGUGUGGCGGAAUUGGCAGACGCACCAGACUUAGGAUCUGGCGCCUU---UGGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAA------UUAAAUAAC ...((--((((..((((((((..........(.((((.((.((((((....).))))).)).))---)).)..(((((.....)))))..))))))))...))))------))....... ( -40.20) >Bcere.0 4650521 109 + 5224283/0-120 AUAAA--UUAAUAUGCGGGUGUGGCGGAAUUGGCAGACGCACCAGACUUAGGAUCUGGCGCCUU---UGGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAA------UUAAAUAAC ...((--((((..((((((((..........(.((((.((.((((((....).))))).)).))---)).)..(((((.....)))))..))))))))...))))------))....... ( -40.20) >Bclau.0 2626639 120 + 4303871/0-120 CUGAGCUUUGGUAUGCGGGUGUGGCGGAAUUGGCAGACGCGCUAGAAUCAGGCUCUAGUGUCCUAACGGACGUGGGGGUUCAAGUCCCUUCACCCGCAUUUCAAAAACUCUUUAAGCAAC ..(((.(((((.(((((((((.((.(((.(((((...(((((((((.......))))))((((....)))))))...).)))).))))).))))))))).)))))..))).......... ( -48.30) >consensus AUAAA__UUAAUAUGCGGGUGUGGCGGAAUUGGCAGACGCACCAGACUUAGGAUCUGGCGCCUU___UGGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAA______UUAAAUAAC .........((..((((((((.((.(((.(((((...(((.(((((.......))))).(((......))))))...).)))).))))).))))))))..)).................. (-38.74 = -36.97 + -1.77)

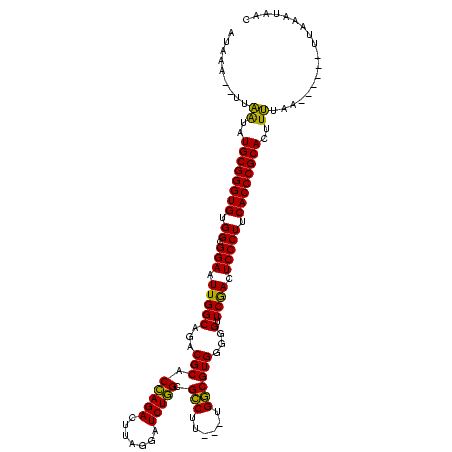
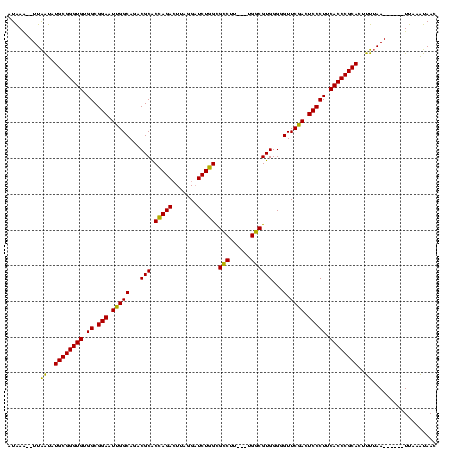
| Location | 4,651,543 – 4,651,652 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -34.05 |
| Energy contribution | -32.83 |
| Covariance contribution | -1.22 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.31 |
| Structure conservation index | 0.86 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4651543 109 + 5227293/2-122 UAUUUAA------UUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCCA---AAGGCGCCAGAUCCUAAGUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUAUUAA--UUUAUUU .....((------((((..(((((((((..(((.(.....).)))..(.((---.(.(((((((((.....))))))))).).)).)..........))))))))).))))--))..... ( -38.90) >Bcere.0 4650521 109 + 5224283/2-122 UAUUUAA------UUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCCA---AAGGCGCCAGAUCCUAAGUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUAUUAA--UUUAUUU .....((------((((..(((((((((..(((.(.....).)))..(.((---.(.(((((((((.....))))))))).).)).)..........))))))))).))))--))..... ( -38.90) >Bclau.0 2626639 120 + 4303871/2-122 UGCUUAAAGAGUUUUUGAAAUGCGGGUGAAGGGACUUGAACCCCCACGUCCGUUAGGACACUAGAGCCUGAUUCUAGCGCGUCUGCCAAUUCCGCCACACCCGCAUACCAAAGCUCAGUU ........(((((((.(..(((((((((..((((.(((.........((((....))))..(((((((((....))).)).)))).))).))).)..)))))))))..)))))))).... ( -41.00) >consensus UAUUUAA______UUAAAAGUGCGGGUGAAGGGAGUCGAACCCCCACGCCA___AAGGCGCCAGAUCCUAAGUCUGGUGCGUCUGCCAAUUCCGCCACACCCGCAUAUUAA__UUUAUUU ...................(((((((((..(((.........)))..(((......)))(((((((.....)))))))(((...........)))..))))))))).............. (-34.05 = -32.83 + -1.22)

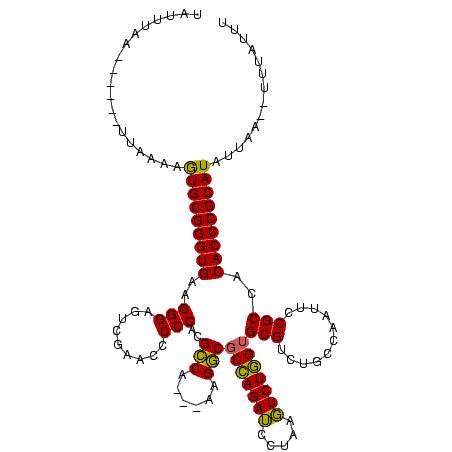
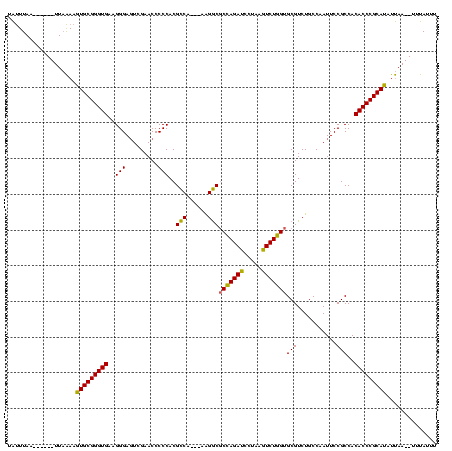
| Location | 4,651,543 – 4,651,652 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.52 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -38.74 |
| Energy contribution | -36.97 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.16 |
| Mean z-score | -3.12 |
| Structure conservation index | 0.90 |
| SVM decision value | 3.10 |
| SVM RNA-class probability | 0.998448 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4651543 109 + 5227293/2-122 AAAUAAA--UUAAUAUGCGGGUGUGGCGGAAUUGGCAGACGCACCAGACUUAGGAUCUGGCGCCUU---UGGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAA------UUAAAUA .....((--((((..((((((((..........(.((((.((.((((((....).))))).)).))---)).)..(((((.....)))))..))))))))...))))------))..... ( -40.20) >Bcere.0 4650521 109 + 5224283/2-122 AAAUAAA--UUAAUAUGCGGGUGUGGCGGAAUUGGCAGACGCACCAGACUUAGGAUCUGGCGCCUU---UGGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAA------UUAAAUA .....((--((((..((((((((..........(.((((.((.((((((....).))))).)).))---)).)..(((((.....)))))..))))))))...))))------))..... ( -40.20) >Bclau.0 2626639 120 + 4303871/2-122 AACUGAGCUUUGGUAUGCGGGUGUGGCGGAAUUGGCAGACGCGCUAGAAUCAGGCUCUAGUGUCCUAACGGACGUGGGGGUUCAAGUCCCUUCACCCGCAUUUCAAAAACUCUUUAAGCA ....(((.(((((.(((((((((.((.(((.(((((...(((((((((.......))))))((((....)))))))...).)))).))))).))))))))).)))))..)))........ ( -48.30) >consensus AAAUAAA__UUAAUAUGCGGGUGUGGCGGAAUUGGCAGACGCACCAGACUUAGGAUCUGGCGCCUU___UGGCGUGGGGGUUCGACUCCCUUCACCCGCACUUUUAA______UUAAAUA ...........((..((((((((.((.(((.(((((...(((.(((((.......))))).(((......))))))...).)))).))))).))))))))..))................ (-38.74 = -36.97 + -1.77)

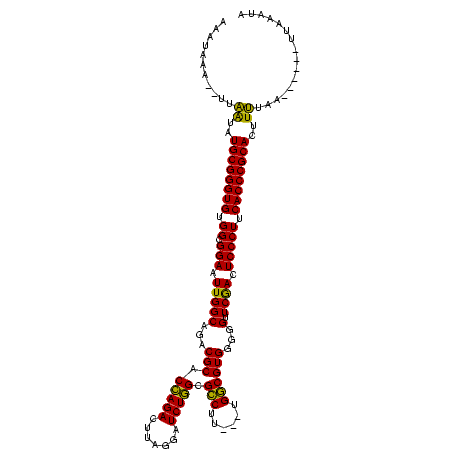
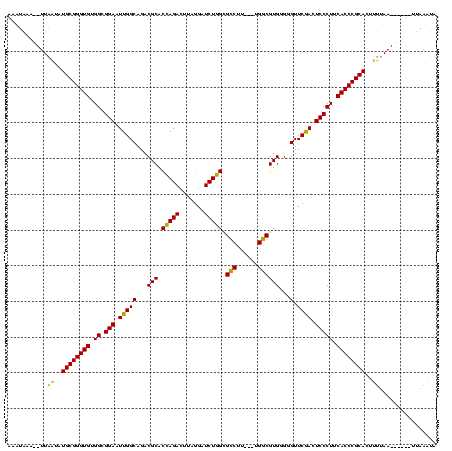
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:19:44 2006