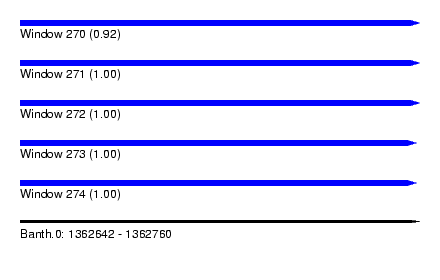
| Sequence ID | Banth.0 |
|---|---|
| Location | 1,362,642 – 1,362,760 |
| Length | 118 |
| Max. P | 0.999999 |

| Location | 1,362,642 – 1,362,760 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.01 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -25.74 |
| Energy contribution | -24.53 |
| Covariance contribution | -1.21 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1362642 118 + 5227293/0-120 AAAUACAAAUACAAAGCUUAUCAAGAGAAGCGGAGGGAACUGGCCCGGCGAAGCUCGGCAACCUGCUUAU--AGAAAGCAAGGUGCUAAAUCCAGCAAAAUGGAAUCCAUUUUGAAAGAU ........................(((...((..(((......)))..))...)))((((.(((((((..--...)))).)))))))..(((...((((((((...))))))))...))) ( -31.00) >Bcere.0 1518502 118 + 5224283/0-120 AAAUACAAAUACAAAGCUUAUCAAGAGAAGCGGAGGGAACUGGCCCGGCGAAGCUCGGCAACCUGCUUAU--AGAAAGCAAGGUGCUAAAUCCAGCAAAAUGGAAUCCAUUUUGAAAGAU ........................(((...((..(((......)))..))...)))((((.(((((((..--...)))).)))))))..(((...((((((((...))))))))...))) ( -31.00) >Bclau.0 1701635 119 + 4303871/0-120 AAAAAUGAAUA-AGAACUUAUCAAGAGUGGCGGAGGGACCUGACCCAAUGAAGCCCGGCAACCAGUUUAUUUGUAAACCACGGUGCUACAUUCAGCAGAGCAGCUUUUGUUCUGAAAGAU .....(((((.-.((.....))....(((((...(((......)))......(((.(....)..(((((....)))))...))))))))))))).((((((((...))))))))...... ( -25.90) >consensus AAAUACAAAUACAAAGCUUAUCAAGAGAAGCGGAGGGAACUGGCCCGGCGAAGCUCGGCAACCUGCUUAU__AGAAAGCAAGGUGCUAAAUCCAGCAAAAUGGAAUCCAUUUUGAAAGAU ........................(((...((..(((......)))..))...)))((((.(((((((.......)))).)))))))..(((...((((((((...))))))))...))) (-25.74 = -24.53 + -1.21)

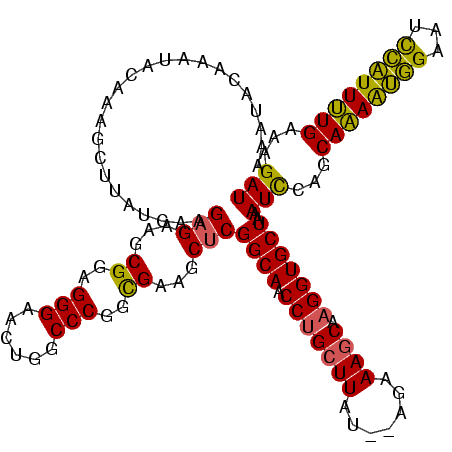
| Location | 1,362,642 – 1,362,760 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -34.47 |
| Consensus MFE | -21.30 |
| Energy contribution | -22.20 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.04 |
| Structure conservation index | 0.62 |
| SVM decision value | 5.00 |
| SVM RNA-class probability | 0.999967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1362642 118 + 5227293/40-160 UGGCCCGGCGAAGCUCGGCAACCUGCUUAU--AGAAAGCAAGGUGCUAAAUCCAGCAAAAUGGAAUCCAUUUUGAAAGAUAAGGUAAAAUAUAUUACCGAACAGUCUUUUCGAAAUGGGA ..(((.(((...))).))).((((((((..--...)))).))))......((((......)))).(((((((((((((((..(((((......))))).....))).)))))))))))). ( -37.60) >Bcere.0 1518502 118 + 5224283/40-160 UGGCCCGGCGAAGCUCGGCAACCUGCUUAU--AGAAAGCAAGGUGCUAAAUCCAGCAAAAUGGAAUCCAUUUUGAAAGAUAAGGUAAAAUAUAUUACCGAACAGUCUUUUCGAAAUGGGA ..(((.(((...))).))).((((((((..--...)))).))))......((((......)))).(((((((((((((((..(((((......))))).....))).)))))))))))). ( -37.60) >Bclau.0 1701635 111 + 4303871/40-160 UGACCCAAUGAAGCCCGGCAACCAGUUUAUUUGUAAACCACGGUGCUACAUUCAGCAGAGCAGCUUUUGUUCUGAAAGAUAAGUGAGG---------CGAACCCCUUUUCUUAUCUGAAA ......((((.((((((....)..(((((....)))))...)).))).))))...((((((((...))))))))..(((((((.((((---------......))))..))))))).... ( -28.20) >consensus UGGCCCGGCGAAGCUCGGCAACCUGCUUAU__AGAAAGCAAGGUGCUAAAUCCAGCAAAAUGGAAUCCAUUUUGAAAGAUAAGGUAAAAUAUAUUACCGAACAGUCUUUUCGAAAUGGGA ...((((.(((((...((((.(((((((.......)))).)))))))..(((...((((((((...))))))))...)))..(((((......))))).........)))))...)))). (-21.30 = -22.20 + 0.90)

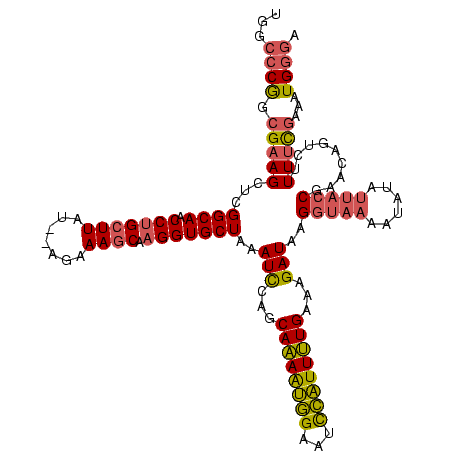
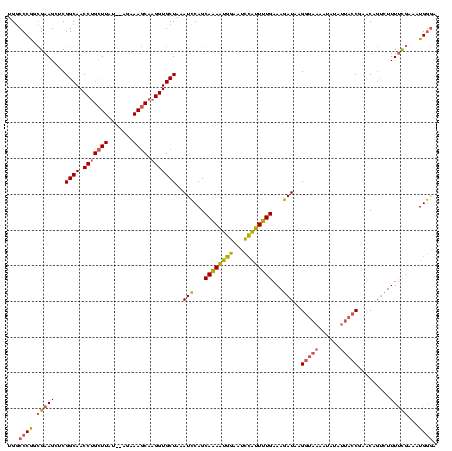
| Location | 1,362,642 – 1,362,760 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.63 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -22.82 |
| Energy contribution | -21.50 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.93 |
| SVM RNA-class probability | 0.999709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1362642 118 + 5227293/40-160 UCCCAUUUCGAAAAGACUGUUCGGUAAUAUAUUUUACCUUAUCUUUCAAAAUGGAUUCCAUUUUGCUGGAUUUAGCACCUUGCUUUCU--AUAAGCAGGUUGCCGAGCUUCGCCGGGCCA .(((....((((......(((((((((......)))))..((((..((((((((...))))))))..))))...(((.(((((((...--..))))))).))).))))))))..)))... ( -34.00) >Bcere.0 1518502 118 + 5224283/40-160 UCCCAUUUCGAAAAGACUGUUCGGUAAUAUAUUUUACCUUAUCUUUCAAAAUGGAUUCCAUUUUGCUGGAUUUAGCACCUUGCUUUCU--AUAAGCAGGUUGCCGAGCUUCGCCGGGCCA .(((....((((......(((((((((......)))))..((((..((((((((...))))))))..))))...(((.(((((((...--..))))))).))).))))))))..)))... ( -34.00) >Bclau.0 1701635 111 + 4303871/40-160 UUUCAGAUAAGAAAAGGGGUUCG---------CCUCACUUAUCUUUCAGAACAAAAGCUGCUCUGCUGAAUGUAGCACCGUGGUUUACAAAUAAACUGGUUGCCGGGCUUCAUUGGGUCA ....(((((((....((((....---------)))).)))))))..((((.((.....)).))))((.((((.(((.((((((((((....))))))).....)))))).)))).))... ( -27.10) >consensus UCCCAUUUCGAAAAGACUGUUCGGUAAUAUAUUUUACCUUAUCUUUCAAAAUGGAUUCCAUUUUGCUGGAUUUAGCACCUUGCUUUCU__AUAAGCAGGUUGCCGAGCUUCGCCGGGCCA .(((...((.....))..(((((.................((((..(((((((.....)))))))..))))...((((((.((((.......))))))).))))))))......)))... (-22.82 = -21.50 + -1.32)

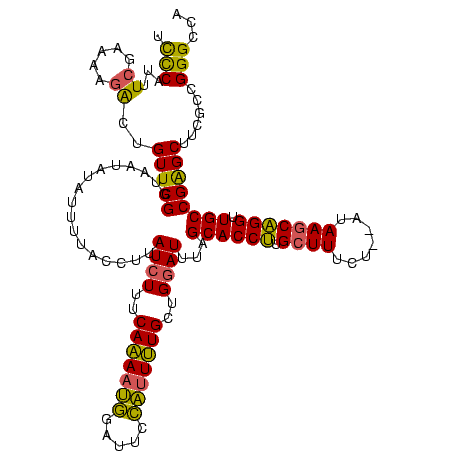
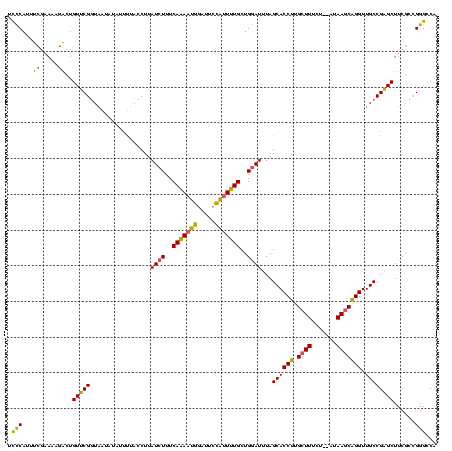
| Location | 1,362,642 – 1,362,759 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.43 |
| Mean single sequence MFE | -32.13 |
| Consensus MFE | -21.29 |
| Energy contribution | -19.87 |
| Covariance contribution | -1.43 |
| Combinations/Pair | 1.31 |
| Mean z-score | -3.65 |
| Structure conservation index | 0.66 |
| SVM decision value | 6.65 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1362642 117 + 5227293/60-180 ACCUGCUUAU--AGAAAGCAAGGUGCUAAAUCCAGCAAAAUGGAAUCCAUUUUGAAAGAUAAGGUAAAAUAUAUUACCGAACAGUCUUUUCGAAAUGGGAAAGAUUUUUUUUA-UGAAUA ((((((((..--...)))).))))......((((......)))).(((((((((((((((..(((((......))))).....))).))))))))))))..............-...... ( -30.20) >Bcere.0 1518502 117 + 5224283/60-180 ACCUGCUUAU--AGAAAGCAAGGUGCUAAAUCCAGCAAAAUGGAAUCCAUUUUGAAAGAUAAGGUAAAAUAUAUUACCGAACAGUCUUUUCGAAAUGGGAAAGAUUUUUUUUA-UGAAUA ((((((((..--...)))).))))......((((......)))).(((((((((((((((..(((((......))))).....))).))))))))))))..............-...... ( -30.20) >Bclau.0 1701635 111 + 4303871/60-180 ACCAGUUUAUUUGUAAACCACGGUGCUACAUUCAGCAGAGCAGCUUUUGUUCUGAAAGAUAAGUGAGG---------CGAACCCCUUUUCUUAUCUGAAAAGGGGUUUUUUCAAUGGAGA .(((.((((((((((.((....))..)))......((((((((...))))))))..)))))))(((((---------.((((((((((((......))))))))))))))))).)))... ( -36.00) >consensus ACCUGCUUAU__AGAAAGCAAGGUGCUAAAUCCAGCAAAAUGGAAUCCAUUUUGAAAGAUAAGGUAAAAUAUAUUACCGAACAGUCUUUUCGAAAUGGGAAAGAUUUUUUUUA_UGAAUA ((((((((.......)))).)))).....(((...((((((((...))))))))...)))..(((((......)))))(((..((((((((......))))))))..))).......... (-21.29 = -19.87 + -1.43)

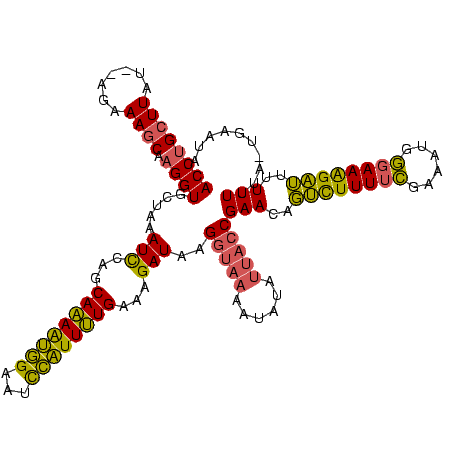
| Location | 1,362,642 – 1,362,759 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.43 |
| Mean single sequence MFE | -25.40 |
| Consensus MFE | -14.22 |
| Energy contribution | -14.00 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.56 |
| SVM decision value | 3.21 |
| SVM RNA-class probability | 0.998747 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 1362642 117 + 5227293/60-180 UAUUCA-UAAAAAAAAUCUUUCCCAUUUCGAAAAGACUGUUCGGUAAUAUAUUUUACCUUAUCUUUCAAAAUGGAUUCCAUUUUGCUGGAUUUAGCACCUUGCUUUCU--AUAAGCAGGU ......-.........(((((.(......).))))).((((.(((((......)))))..((((..((((((((...))))))))..))))..)))).(((((((...--..))))))). ( -23.60) >Bcere.0 1518502 117 + 5224283/60-180 UAUUCA-UAAAAAAAAUCUUUCCCAUUUCGAAAAGACUGUUCGGUAAUAUAUUUUACCUUAUCUUUCAAAAUGGAUUCCAUUUUGCUGGAUUUAGCACCUUGCUUUCU--AUAAGCAGGU ......-.........(((((.(......).))))).((((.(((((......)))))..((((..((((((((...))))))))..))))..)))).(((((((...--..))))))). ( -23.60) >Bclau.0 1701635 111 + 4303871/60-180 UCUCCAUUGAAAAAACCCCUUUUCAGAUAAGAAAAGGGGUUCG---------CCUCACUUAUCUUUCAGAACAAAAGCUGCUCUGCUGAAUGUAGCACCGUGGUUUACAAAUAAACUGGU ...(((.(((...(((((((((((......)))))))))))..---------..))).................((((..(..(((((....)))))..)..))))..........))). ( -29.00) >consensus UAUUCA_UAAAAAAAAUCUUUCCCAUUUCGAAAAGACUGUUCGGUAAUAUAUUUUACCUUAUCUUUCAAAAUGGAUUCCAUUUUGCUGGAUUUAGCACCUUGCUUUCU__AUAAGCAGGU ......................(((.................(((((......)))))........(((((((.....))))))).))).......((((.((((.......)))))))) (-14.22 = -14.00 + -0.22)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:19:17 2006