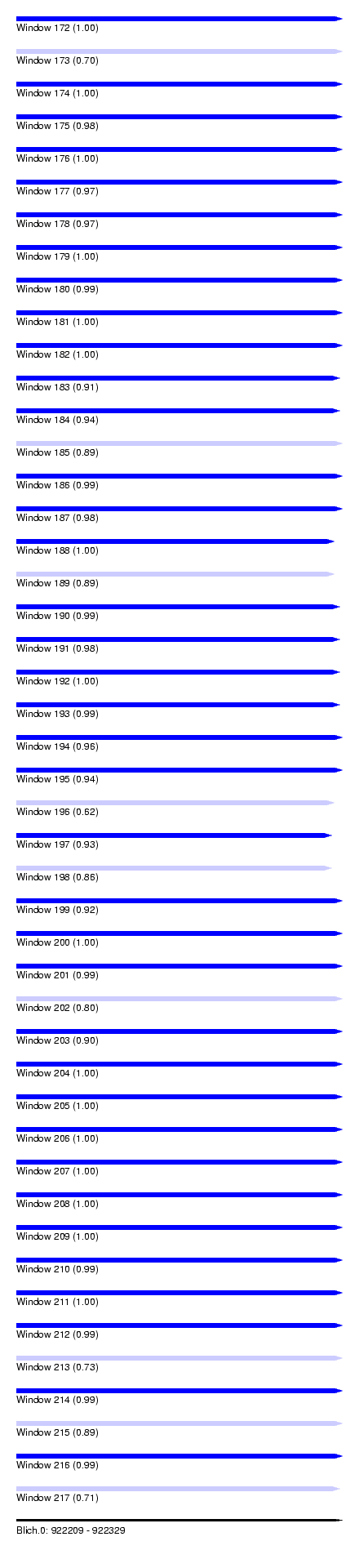
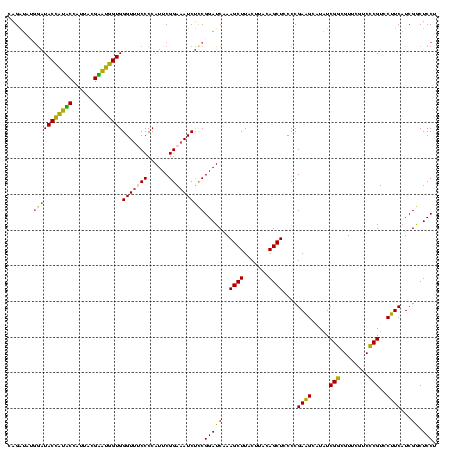
| Sequence ID | Blich.0 |
|---|---|
| Location | 922,209 – 922,329 |
| Length | 120 |
| Max. P | 0.999995 |

| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.78 |
| Mean single sequence MFE | -43.23 |
| Consensus MFE | -42.43 |
| Energy contribution | -40.02 |
| Covariance contribution | -2.41 |
| Combinations/Pair | 1.28 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
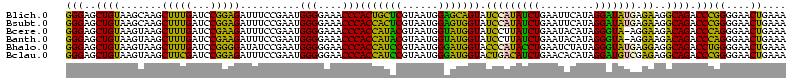
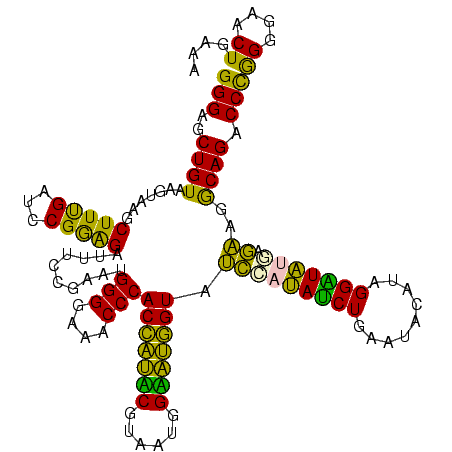
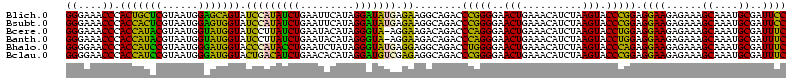
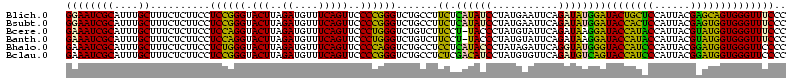
>Blich.0 922209 120 + 4222334/40-160 AGGAGCCGAUGAAGGACGGGACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUG .(((...(((.((((.(((........)))...((((((((....))).)))))..)))).)))(((......)))...(((....)))(((((((......))))))).)))....... ( -43.60) >Bsubt.0 947750 120 + 4214630/40-160 AGGAGCCGAUGAAGGACGGGACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUG .(((...(((.((((.(((........)))...((((((((....))).)))))..)))).)))(((......)))...(((....)))(((((((......))))))).)))....... ( -43.60) >Bcere.0 606627 120 + 5224283/40-160 AGGAGUCGAUGAAGGACGGGACUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUG .......(((((.(((((((((....)........((((((((((((.......)))))...)))))))..)))))...(((....)))(((((((......))))))).)))))))).. ( -42.10) >Banth.0 534125 120 + 5227293/40-160 AGGAGUCGAUGAAGGACGGGACUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUG .......(((((.(((((((((....)........((((((((((((.......)))))...)))))))..)))))...(((....)))(((((((......))))))).)))))))).. ( -42.10) >Bhalo.0 119923 120 + 4202352/40-160 AGGAGCCGAAGAAGGACGCGACGAACGGCGAAACGCCUCGGGGAGCUGUAAGUAAGCUUUGAUCCGGGGAUAUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACCCAUACCUG .(((((((..(..(....)..)...))))......((((((((((((.......)))))...)))))))...)))..(((((((...))(((((((......))))))).)))))..... ( -42.90) >Bclau.0 935920 120 + 4303871/40-160 AGGAGCCGAUGAAGGACGCGACGAACCGCGAUAGGCUUCGGGGAGCUGUAAGUAAGCUUCGAUCCGGAGAUUUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACUGACAUCUG .......((((..(((((((......)))).....((((((((((((.......))))))...))))))...)))....(((....)))(((((((......))))))).....)))).. ( -45.10) >consensus AGGAGCCGAUGAAGGACGGGACGAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGAAUGGUAUCCAUAUCUG .......((((..((((((........))).....((((((((((((.......)))))...)))))))...)))....(((....)))(((((((......))))))).....)))).. (-42.43 = -40.02 + -2.41)

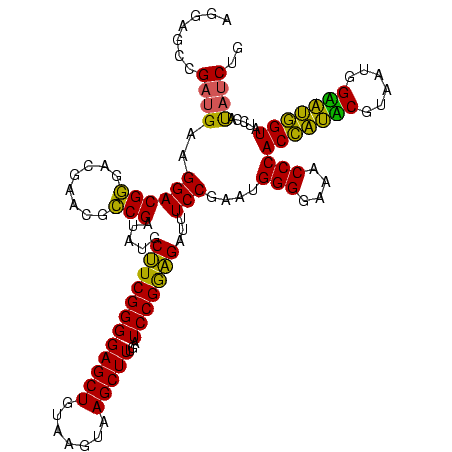
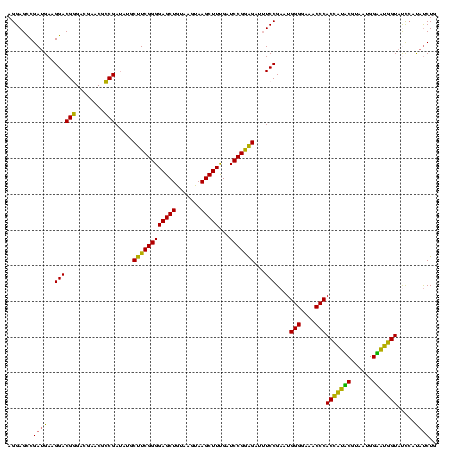
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.78 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -30.74 |
| Energy contribution | -28.58 |
| Covariance contribution | -2.16 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/40-160 CAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUUCGUCCCGUCCUUCAUCGGCUCCU ..(((.((((((((((((......))))))))(((((((......))))))))))).)))..((((.((.....)).....((((.....(((........)))..))))...))))... ( -33.90) >Bsubt.0 947750 120 + 4214630/40-160 CAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUUCGUCCCGUCCUUCAUCGGCUCCU ..(((.((((((((((((......))))))))(((((((......))))))))))).)))..((((.((.....)).....((((.....(((........)))..))))...))))... ( -33.90) >Bcere.0 606627 120 + 5224283/40-160 CAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUAGUCCCGUCCUUCAUCGACUCCU ..((((((((((((((((......)))))))((((((((......))))).((((((.....((((.......))))..))))))...............))))))))).)))....... ( -33.70) >Banth.0 534125 120 + 5227293/40-160 CAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUAGUCCCGUCCUUCAUCGACUCCU ..((((((((((((((((......)))))))((((((((......))))).((((((.....((((.......))))..))))))...............))))))))).)))....... ( -33.70) >Bhalo.0 119923 120 + 4202352/40-160 CAGGUAUGGGUACCAUCCCAUUACGGAUGGUGGGUUCCCCCAUUCGGAUAUCCCCGGAUCAAAGCUUACUUACAGCUCCCCGAGGCGUUUCGCCGUUCGUCGCGUCCUUCUUCGGCUCCU ..((.(((((......)))))..(((((((.((....))))))))).......))(((....((((.......))))..((((((.....(((........))).....)))))).))). ( -36.10) >Bclau.0 935920 120 + 4303871/40-160 CAGAUGUCAGUACCAUCCCAUUACGGAUGGUGGGUUCCCCCAUUCGGAAAUCUCCGGAUCGAAGCUUACUUACAGCUCCCCGAAGCCUAUCGCGGUUCGUCGCGUCCUUCAUCGGCUCCU ..((.(((...(((((((......)))))))(((.....(.(((((((....))))))).).((((.......)))).)))((((.....(((((....)))))..))))...))))).. ( -35.40) >consensus CAGAUAUGGAUACCAUACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUCGUCCCGUCCUUCAUCGGCUCCU .......(((((((((((......))))))))(((((((......))))))))))(((((..((((.......))))....((((.....(((........)))..))))...)).))). (-30.74 = -28.58 + -2.16)

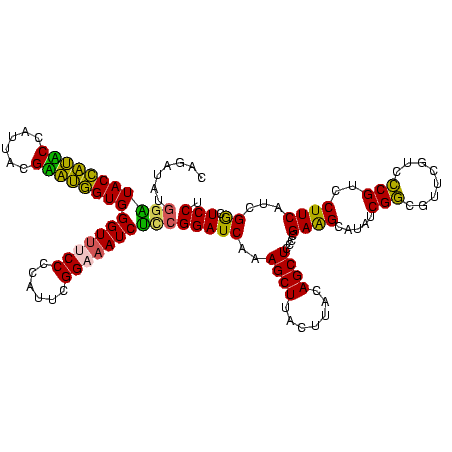
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -41.00 |
| Consensus MFE | -37.29 |
| Energy contribution | -33.67 |
| Covariance contribution | -3.63 |
| Combinations/Pair | 1.52 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/80-200 GGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAA .((((((.......))))))..(((((.(.(((((....(((....)))(((((((......))))))).(((((((((.........))))))).)).)).)))))))))......... ( -41.30) >Bsubt.0 947750 120 + 4214630/80-200 GGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAA .((((((.......))))))..(((((.(.(((((....(((....)))(((((((......))))))).(((((((((.........))))))).)).)).)))))))))......... ( -41.30) >Bcere.0 606627 119 + 5224283/80-200 GGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAA (((..((((.......(((((...)))))..........(((....)))(((((((......))))))).(((((((((.........)))))-))))..)))).)))((....)).... ( -37.50) >Banth.0 534125 119 + 5227293/80-200 GGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAA (((..((((.......(((((...)))))..........(((....)))(((((((......))))))).(((((((((.........)))))-))))..)))).)))((....)).... ( -37.50) >Bhalo.0 119923 120 + 4202352/80-200 GGGAGCUGUAAGUAAGCUUUGAUCCGGGGAUAUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACCCAUACCUGAAUCUAUAGGGUAUGAGGAGGCAGACCUGGGGAACUGAAA .((((((.......))))))..((((((.....((....(((....)))(((((((......))))))).(((((((((.........))))))).)).))....))))))......... ( -44.70) >Bclau.0 935920 120 + 4303871/80-200 GGGAGCUGUAAGUAAGCUUCGAUCCGGAGAUUUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACUGACAUCUGAACACAUAGGAUGUCGAGAGGCAGACCCGGGGAACUGAAA .((((((.......))))))..(((((.(.(((((....(((....)))(((((((......))))))).(((((((((.........))))))).)).)).)))))))))......... ( -43.70) >consensus GGGAGCUGUAAGUAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAA (((..((((.......(((((...)))))..........(((....)))(((((((......))))))).(((((((((.........))))))).))..)))).)))((....)).... (-37.29 = -33.67 + -3.63)
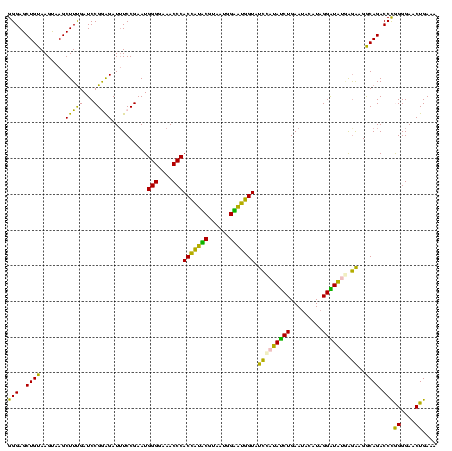
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.10 |
| Mean single sequence MFE | -32.88 |
| Consensus MFE | -29.33 |
| Energy contribution | -27.08 |
| Covariance contribution | -2.24 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/80-200 UUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCC ............(((...(((.((.((((((...((....))))))))))((((((((......)))))))))))...)))(((((((....)))))))...((((.......))))... ( -34.00) >Bsubt.0 947750 120 + 4214630/80-200 UUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCC ............(((...(((.((.((((((...((....))))))))))((((((((......)))))))))))...)))(((((((....)))))))...((((.......))))... ( -34.00) >Bcere.0 606627 119 + 5224283/80-200 UUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCC ...(((....)))((((((...((((-((..((........)).))))))((((((((......))))))))(((((((......)))))))..))))))..((((.......))))... ( -29.90) >Banth.0 534125 119 + 5227293/80-200 UUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCC ...(((....)))((((((...((((-((..((........)).))))))((((((((......))))))))(((((((......)))))))..))))))..((((.......))))... ( -29.90) >Bhalo.0 119923 120 + 4202352/80-200 UUUCAGUUCCCCAGGUCUGCCUCCUCAUACCCUAUAGAUUCAGGUAUGGGUACCAUCCCAUUACGGAUGGUGGGUUCCCCCAUUCGGAUAUCCCCGGAUCAAAGCUUACUUACAGCUCCC .............((((((....((((((((...........)))))))).(((((((......)))))))(((.(((.......)))...)))))))))..((((.......))))... ( -33.40) >Bclau.0 935920 120 + 4303871/80-200 UUUCAGUUCCCCGGGUCUGCCUCUCGACAUCCUAUGUGUUCAGAUGUCAGUACCAUCCCAUUACGGAUGGUGGGUUCCCCCAUUCGGAAAUCUCCGGAUCGAAGCUUACUUACAGCUCCC .(((........(((...(((....((((((...........))))))..((((((((......)))))))))))...)))(((((((....))))))).)))(((.......))).... ( -36.10) >consensus UUUCAGUUCCCCGGGUCUGCCUUCCCAUACCCUAUGUAUUCAGAUAUGGAUACCAUACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUACUUACAGCUCCC .............((((((...((.((((((...........)))))))).(((((((......)))))))((((((((......)))))))).))))))..((((.......))))... (-29.33 = -27.08 + -2.24)

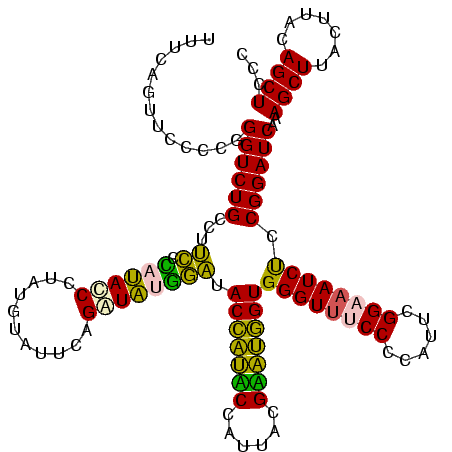
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -34.81 |
| Energy contribution | -31.32 |
| Covariance contribution | -3.49 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.77 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.13 |
| SVM RNA-class probability | 0.998530 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/120-240 GGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUCC ((....)).(((((((......))))))).(((...((((..(((.((.....)).)))..)))).(((((..(((..........))).))))).)))...(((.((....))..))). ( -37.80) >Bsubt.0 947750 120 + 4214630/120-240 GGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUCC ((....)).(((((((......))))))).(((...((((..(((.((.....)).)))..)))).(((((..(((..........))).))))).)))...(((.((....))..))). ( -37.80) >Bcere.0 606627 119 + 5224283/120-240 GGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAGAGAAAGCAAAUGCGAUUUC ((....)).(((((((......))))))).(((((((((.........)))))-))))........(((((..(((..........))).))))).....((((..((....))..)))) ( -35.50) >Banth.0 534125 119 + 5227293/120-240 GGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAGAGAAAGCAAAUGCGAUUUC ((....)).(((((((......))))))).(((((((((.........)))))-))))........(((((..(((..........))).))))).....((((..((....))..)))) ( -35.50) >Bhalo.0 119923 120 + 4202352/120-240 GGGGAACCCACCAUCCGUAAUGGGAUGGUACCCAUACCUGAAUCUAUAGGGUAUGAGGAGGCAGACCUGGGGAACUGAAACAUCUAAGUACCCAGAGGAAGAGAAAGCAAAUGCGAUUUC (((...)))(((((((......))))))).(((((((((.........))))))).)).......(((.(((.(((..........))).)))..)))..((((..((....))..)))) ( -37.80) >Bclau.0 935920 120 + 4303871/120-240 GGGGAACCCACCAUCCGUAAUGGGAUGGUACUGACAUCUGAACACAUAGGAUGUCGAGAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUUC (((....(((((((((......))))))).(((((((((.........))))))).)).))....))).(((.(((..........))).))).......((((..((....))..)))) ( -37.00) >consensus GGGAAACCCACCAUACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUUC ((....)).(((((((......))))))).(((((((((.........))))))).))........(((((..(((..........))).))))).((((......((....))..)))) (-34.81 = -31.32 + -3.49)
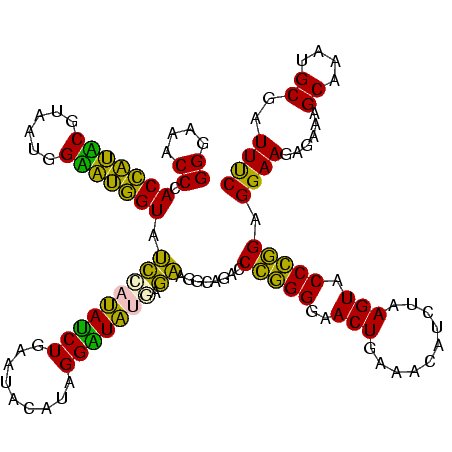
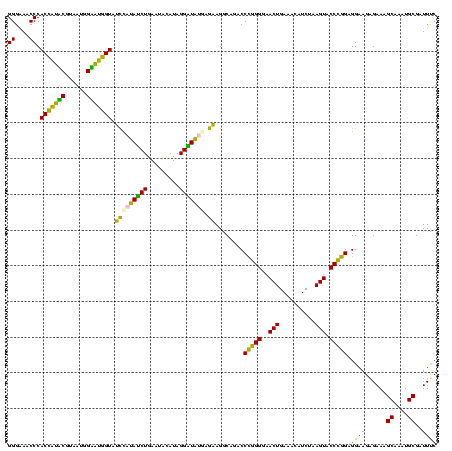
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.66 |
| Mean single sequence MFE | -32.77 |
| Consensus MFE | -30.18 |
| Energy contribution | -27.08 |
| Covariance contribution | -3.10 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/120-240 GGAAUCGCAUUUGCUUUCUCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCC ((((((((....)).......(((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..)))((((((((......)))))))))))))).. ( -35.00) >Bsubt.0 947750 120 + 4214630/120-240 GGAAUCGCAUUUGCUUUCUCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCC ((((((((....)).......(((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..)))((((((((......)))))))))))))).. ( -35.00) >Bcere.0 606627 119 + 5224283/120-240 GAAAUCGCAUUUGCUUUCUCUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCC ((((((((....)).......(((((((((.(((..((....)))))..)))))(((((.......-(((.....)))..))))).))))((((((((......)))))))))))))).. ( -30.10) >Banth.0 534125 119 + 5227293/120-240 GAAAUCGCAUUUGCUUUCUCUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCC ((((((((....)).......(((((((((.(((..((....)))))..)))))(((((.......-(((.....)))..))))).))))((((((((......)))))))))))))).. ( -30.10) >Bhalo.0 119923 120 + 4202352/120-240 GAAAUCGCAUUUGCUUUCUCUUCCUCUGGGUACUUAGAUGUUUCAGUUCCCCAGGUCUGCCUCCUCAUACCCUAUAGAUUCAGGUAUGGGUACCAUCCCAUUACGGAUGGUGGGUUCCCC ((((..((....))))))......((((((.(((..((....)))))..))))))...(((..((((((((...........))))))))((((((((......)))))))))))..... ( -33.10) >Bclau.0 935920 120 + 4303871/120-240 GAAAUCGCAUUUGCUUUCUCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUCUCGACAUCCUAUGUGUUCAGAUGUCAGUACCAUCCCAUUACGGAUGGUGGGUUCCCC ((((..((....)))))).........(((.(((..((....))))).))).(((...(((....((((((...........))))))..((((((((......))))))))))).))). ( -33.30) >consensus GAAAUCGCAUUUGCUUUCUCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCCCAUACCCUAUGUAUUCAGAUAUGGAUACCAUACCAUUACGAAUGGUGGGUUUCCC ((((((((....))..........((((((.(((..((....)))))..)))))).......((.((((((...........))))))))((((((((......)))))))))))))).. (-30.18 = -27.08 + -3.10)
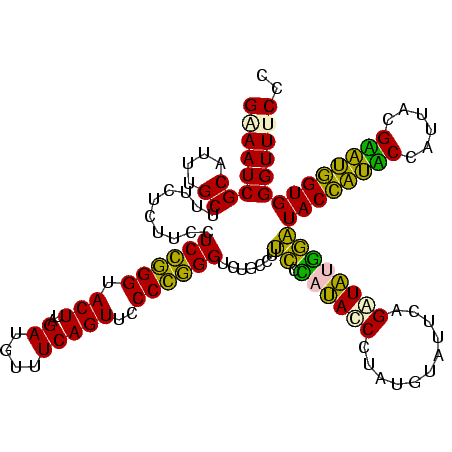
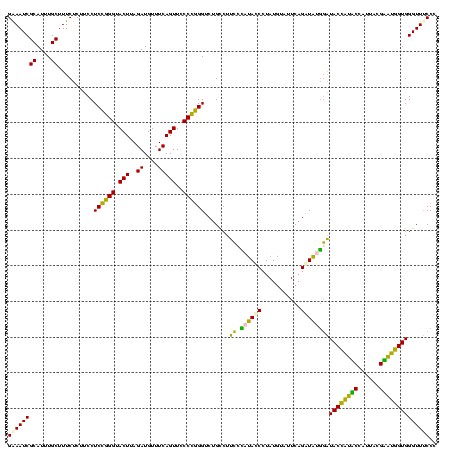
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -38.37 |
| Consensus MFE | -35.79 |
| Energy contribution | -36.32 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.969326 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/440-560 AGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGGUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCGGAUUCCGACGGAAU (((((((((..((..((((....)))).(((........((((((...(((((((((.....)))))).))))))))).....)))..))..))).)))))).(((........)))... ( -39.72) >Bsubt.0 947750 120 + 4214630/440-560 AGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGGUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCGGAUUCCGACGGAAU (((((((((..((..((((....)))).(((........((((((...(((((((((.....)))))).))))))))).....)))..))..))).)))))).(((........)))... ( -39.72) >Bcere.0 606627 120 + 5224283/440-560 AGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGAUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCGGAUUCCGACGGAAU (((((((((..((..((((....)))).(((........((((((...(((..(((....)))...)))...)))))).....)))..))..))).)))))).(((........)))... ( -35.32) >Banth.0 534125 120 + 5227293/440-560 AGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGAUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCAGAUUCCGACGGAAU (((((((((..((..((((....)))).(((........((((((...(((..(((....)))...)))...)))))).....)))..))..))).)))))).....(((((...))))) ( -33.32) >Bhalo.0 119923 120 + 4202352/440-560 AGGAUCUUUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGGUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCAGCUUCCGACGGGGU .(((.....)))...((((.((.((((((((........((((((...(((((((((.....)))))).))))))))).....)))..((((((......))))))))))).)).)))). ( -39.12) >Bclau.0 935920 120 + 4303871/440-560 AGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGGUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCGGAUUCCGACGGGGU (((((((((..((..((((....)))).(((........((((((...(((((((((.....)))))).))))))))).....)))..))..))).))))))((((........)))).. ( -43.02) >consensus AGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGGUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCGGAUUCCGACGGAAU (((((((((..((..((((....)))).(((........((((((...(((((((((.....)))))).))))))))).....)))..))..))).)))))).(((........)))... (-35.79 = -36.32 + 0.53)
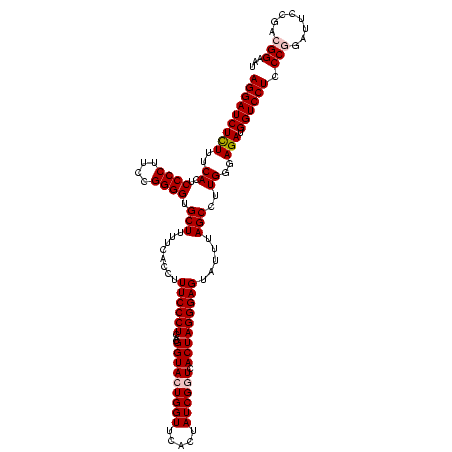
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.39 |
| Mean single sequence MFE | -38.79 |
| Consensus MFE | -38.46 |
| Energy contribution | -38.30 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/520-640 AAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGA ...((((...((.((((....))))....................((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....))))))...)))).. ( -39.80) >Bsubt.0 947750 120 + 4214630/520-640 AAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGA ...((((...((.((((....))))....................((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....))))))...)))).. ( -39.80) >Bcere.0 606627 120 + 5224283/520-640 AAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGA ...((((...((.((((....))))....................((.((..(((((((..((((..(((((....)))))..))))......))))))).....))))))...)))).. ( -39.40) >Banth.0 534125 120 + 5227293/520-640 AAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGA ...((((...((.((((....))))....................((.((..(((((((..((((..(((((....)))))..))))......))))))).....))))))...)))).. ( -39.40) >Bhalo.0 119923 120 + 4202352/520-640 AAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAAAGAUCCUGAAACCGUGUGCCUACAACUAGUUGGAGCCCAUUAACGGGUGACAGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGA ...((((...((.((((....))))....................((.((..((((((...((((..((((......))))..)))).......)))))).....))))))...)))).. ( -35.50) >Bclau.0 935920 120 + 4303871/520-640 AAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAACUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGG .(((......((.((((....)))).)).........)))....(((((((((........((((..(((((....)))))..)))).(((.....))).........))))...))))) ( -38.86) >consensus AAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGA ...((((...((.((((....))))....................((.((..(((((((..((((..(((((....)))))..))))......))))))).....))))))...)))).. (-38.46 = -38.30 + -0.16)

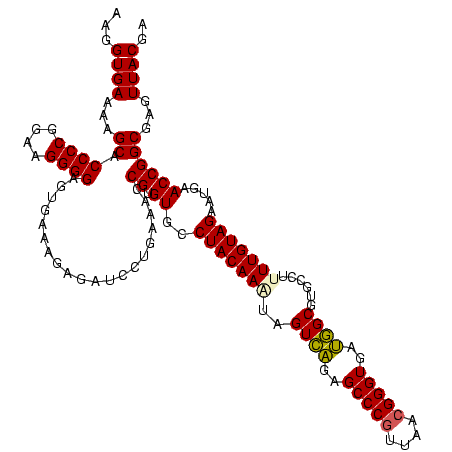
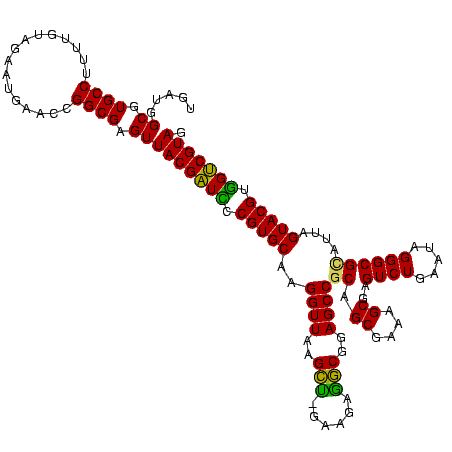
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.23 |
| Mean single sequence MFE | -40.39 |
| Consensus MFE | -39.17 |
| Energy contribution | -37.67 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/600-720 UGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGUUUGAAAAGACGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCAUGAGUACGUGGUCGUAG .....((.((((................)))).))(((((((.(((((..((((..((((....))))..))))((.((....))..((((.....))))))....))))).))))))). ( -41.09) >Bsubt.0 947750 119 + 4214630/600-720 UGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGCA-GAAGAUGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCAUGAGUACGUGGUCGUAG .....((.((((................)))).))(((((((.(((((..((((..(((-.....)))..))))((.((....))..((((.....))))))....))))).))))))). ( -40.69) >Bcere.0 606627 118 + 5224283/600-720 UGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGU-UUAGUACGUGGUCGUAG ((.((((.((((((((.(((....((((..((...((((....)))))).))))...))-)))))))))..))))))((((..(((...((((((.....))-))))..)))..)))).. ( -38.20) >Banth.0 534125 118 + 5227293/600-720 UGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGU-UUAGUACGUGGUCGUAG ((.((((.((((((((.(((....((((..((...((((....)))))).))))...))-)))))))))..))))))((((..(((...((((((.....))-))))..)))..)))).. ( -38.20) >Bhalo.0 119923 119 + 4202352/600-720 UGACAGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUGUCGUGCAAGGUUAAGCU-GAAAAAGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGAAAUAGUACGCCGUCGUAG .....((.((((................)))).))(((((((.(((((..((((..(((-.....)))..))))...((....))..((((.....))))......))))).))))))). ( -37.69) >Bclau.0 935920 119 + 4303871/600-720 UGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGGUAUCGUGCAAGGUUAAGCU-GAUGAAGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCAUUAGUACGAUGCCGUAG .....((.((((................)))).))(((((((((((((..((((..(((-.....)))..))))((.((....))..((((.....))))))....))))))))))))). ( -46.49) >consensus UGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGCU_GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCAUUAGUACGUGGUCGUAG .....((.((((................)))).))(((((((.(((((..((((..(((......)))..))))((.((....))..((((.....))))))....))))).))))))). (-39.17 = -37.67 + -1.50)


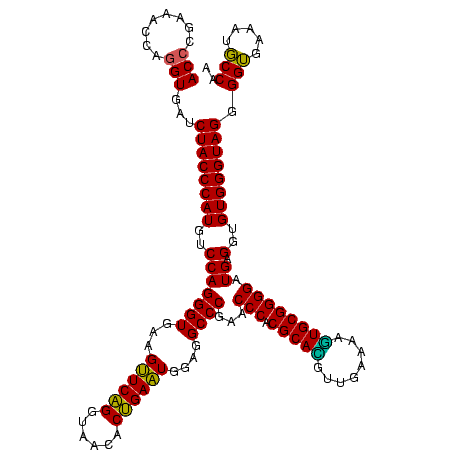
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -49.43 |
| Consensus MFE | -48.85 |
| Energy contribution | -47.47 |
| Covariance contribution | -1.38 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.88 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.90 |
| SVM RNA-class probability | 0.999995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/720-840 ACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAA (((.(....).)))..(((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))))(((.....))).. ( -49.70) >Bsubt.0 947750 120 + 4214630/720-840 ACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAA (((.(....).)))..(((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))))(((.....))).. ( -49.70) >Bcere.0 606627 120 + 5224283/720-840 ACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAA (((.(....).)))...((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))).((((...)))).. ( -47.60) >Banth.0 534125 120 + 5227293/720-840 ACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCAUGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAA (((.(....).)))...((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))).((((...)))).. ( -45.40) >Bhalo.0 119923 120 + 4202352/720-840 ACCCGAAACCGUGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCAUGUUGAAAAAUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAA (((((....).......((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))))))).......... ( -47.30) >Bclau.0 935920 120 + 4303871/720-840 ACCCGAAACCGGGUGAUCUACCCAUGUCCAGGGUGAAGGUCGGGUAACACCGACUGGAGGCCCGAACCCACGCAAGUUGAAAAUUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAA (((((....)))))..(((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))))(((.....))).. ( -56.90) >consensus ACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAA (((........)))...((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))).(((.....))).. (-48.85 = -47.47 + -1.38)

| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.72 |
| Mean single sequence MFE | -39.37 |
| Consensus MFE | -38.91 |
| Energy contribution | -38.08 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.28 |
| SVM RNA-class probability | 0.999982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/720-840 UUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGU ..............(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))) ( -39.30) >Bsubt.0 947750 120 + 4214630/720-840 UUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGU ..............(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))) ( -39.30) >Bcere.0 606627 120 + 5224283/720-840 UUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGU ..(((.....))).(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))) ( -41.50) >Banth.0 534125 120 + 5227293/720-840 UUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACAUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGU ..(((.....))).(((((((.........((((((..........)))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))) ( -37.10) >Bhalo.0 119923 120 + 4202352/720-840 UUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCAUUUUUCAACAUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACACGGUUUCGGGU ..........(((((((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))(((....)))...)))) ( -36.40) >Bclau.0 935920 120 + 4303871/720-840 UUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCAAUUUUCAACUUGCGUGGGUUCGGGCCUCCAGUCGGUGUUACCCGACCUUCACCCUGGACAUGGGUAGAUCACCCGGUUUCGGGU ..............(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))) ( -42.60) >consensus UUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGU ..............(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))) (-38.91 = -38.08 + -0.83)

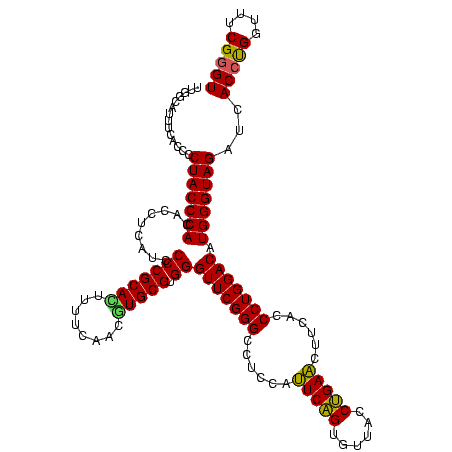
| Location | 922,209 – 922,328 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.16 |
| Mean single sequence MFE | -38.63 |
| Consensus MFE | -33.51 |
| Energy contribution | -33.73 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 119 + 4222334/800-920 AAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAG-GUAAGAGUCUUGGAGGUAGAGCACU ..((((((((((...(((...((((.........)))).)))(((((.((......))))))))))))....((.((((((((((.(((....)-))...)))))))))).))..))))) ( -40.30) >Bsubt.0 947750 119 + 4214630/800-920 AAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAG-GUAAGAGUCUUGGAGGUAGAGCACU ..((((((((((...(((...((((.........)))).)))(((((.((......))))))))))))....((.((((((((((.(((....)-))...)))))))))).))..))))) ( -40.30) >Bcere.0 606627 120 + 5224283/800-920 AAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAAGAGUCUUGGAGGUAGAGCACU ..((((((((((.........(((.(.(((....)))).)))(((((.((......))))))))))))....((.((((((((((...((((....)))))))))))))).))..))))) ( -37.90) >Banth.0 534125 120 + 5227293/800-920 AAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAAGAGUCUUGGAGGUAGAGCACU ..((((((((((.........(((.(.(((....)))).)))(((((.((......))))))))))))....((.((((((((((...((((....)))))))))))))).))..))))) ( -37.90) >Bhalo.0 119923 119 + 4202352/800-920 AAAAUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACUCGGAAAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUCGAGGG-AAGAGUCUUGGAGGUAGAGCACU ....((((((((...(((...((((.........)))).)))((((.(((......))))))))))))....((.((((((((((..((.....))-...)))))))))).))..))).. ( -39.30) >Bclau.0 935920 119 + 4303871/800-920 AAAUUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACUCGGAAAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUCGAGUG-AUGAGUCUUGGAGGUAGAGCACU ....((((((((...(((...((((.........)))).)))((((.(((......))))))))))))....(((((....)))))(((((.((.(-(....)))).)))))...))).. ( -36.10) >consensus AAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUCAAGUGUAAGAGUCUUGGAGGUAGAGCACU ..((((((((((...(((...((((.........)))).)))(((((.((......))))))))))))....((.((((((((((...(......)....)))))))))).))..))))) (-33.51 = -33.73 + 0.22)
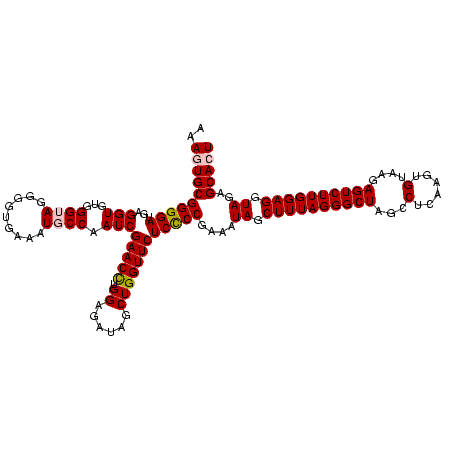
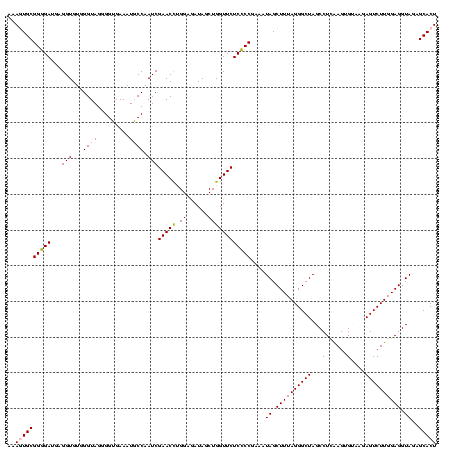
| Location | 922,209 – 922,328 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.99 |
| Mean single sequence MFE | -39.32 |
| Consensus MFE | -37.13 |
| Energy contribution | -35.97 |
| Covariance contribution | -1.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 119 + 4222334/880-1000 GGGCUAGCCUCAAG-GUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGCCCCUACCGGGUUACCGAAUUCAGUCAAACUCCGAAUGCCAAAGACUUAUCCUUGGGAGUCAGACU .((((..(((((((-(...(((((((...(((((((...((((((((...(((((((.....))))).))...))))))))..)))....)))).)))))))..))))))).)..)).)) ( -42.50) >Bsubt.0 947750 119 + 4214630/880-1000 GGGCUAGCCUCAAG-GUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGCCCCUACCGGGUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUGGGAGUCAGACU .((((..(((((((-(...(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))).)..)).)) ( -41.70) >Bcere.0 606627 120 + 5224283/880-1000 GGGCUAGCCUUAAGUGUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCUCAUCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCAGACU .((((..(((((((.(...(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))).)..)).)) ( -34.70) >Banth.0 534125 120 + 5227293/880-1000 GGGCUAGCCUUAAGUGUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCUCAUCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCAGACU .((((..(((((((.(...(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))).)..)).)) ( -34.70) >Bhalo.0 119923 119 + 4202352/880-1000 GGGCUAGCCUCGAGGG-AAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCCCACCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUCGGGAGUCAGACU .((((..(((((((((-..(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).))))).)))))))).)..)).)) ( -41.40) >Bclau.0 935920 119 + 4303871/880-1000 GGGCUAGCCUCGAGUG-AUGAGUCUUGGAGGUAGAGCACUGAUUGGACGAGGGGUCCCCACAGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUUAACUCGGGAGUCAGACU .((((..((((((((.-..(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))).)..)).)) ( -40.90) >consensus GGGCUAGCCUCAAGUGUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCCCACCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUGGGAGUCAGACU .((((..(((((((.(...(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))).)..)).)) (-37.13 = -35.97 + -1.16)


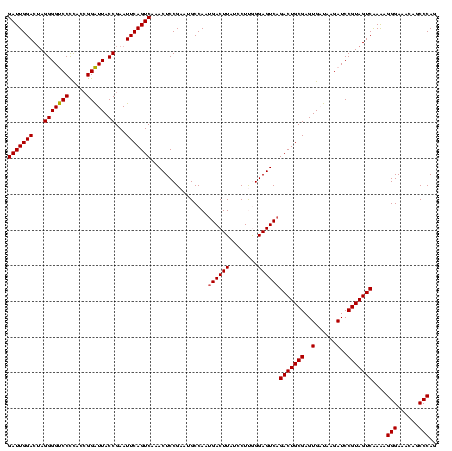
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -36.40 |
| Consensus MFE | -34.39 |
| Energy contribution | -34.12 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893454 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/920-1040 GAUUGGACUAGGGGCCCCUACCGGGUUACCGAAUUCAGUCAAACUCCGAAUGCCAAAGACUUAUCCUUGGGAGUCAGACUGCGAGUGAUAAGAUCCGUAGUCGAAAGGGAAACAGCCCAG (((((((...(((((((.....))))).))...))))))).......((.(.((((.((....)).)))).).)).(((((((..(......)..)))))))....(((......))).. ( -35.90) >Bsubt.0 947750 120 + 4214630/920-1040 GAUUGGACUAGGGGCCCCUACCGGGUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUGGGAGUCAGACUGCGAGUGAUAAGAUCCGUAGUCGAAAGGGAAACAGCCCAG (((((((...(((((((.....))))).))...))))))).......((.(.((((.((....)).)))).).)).(((((((..(......)..)))))))....(((......))).. ( -35.90) >Bcere.0 606627 120 + 5224283/920-1040 GAUUGGACUAGGGGUCCUCAUCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCAGACUGCGAGUGAUAAGAUCCGUAGUCAAAAGGGAAACAGCCCAG .....(((((.((((((((.((((....))))...(((((..(((((......................)))))..))))).)))......))))).)))))....(((......))).. ( -37.95) >Banth.0 534125 120 + 5227293/920-1040 GAUUGGACUAGGGGUCCUCAUCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCAGACUGCGAGUGAUAAGAUCCGUAGUCAAAAGGGAAACAGCCCAG .....(((((.((((((((.((((....))))...(((((..(((((......................)))))..))))).)))......))))).)))))....(((......))).. ( -37.95) >Bhalo.0 119923 120 + 4202352/920-1040 GAUUGGACUAGGGGUCCCCACCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUCGGGAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCAAGAGGGAAACAGCCCAG .....(((((.(((((..((((((....)))....(((((..(((((((.................)).)))))..)))))...)))....))))).)))))....(((......))).. ( -36.13) >Bclau.0 935920 120 + 4303871/920-1040 GAUUGGACGAGGGGUCCCCACAGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUUAACUCGGGAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCAAGAGGGAAACAGCCCAG (((((((...(((((((.....))))).))...)))))))................(((((((......)))))))(((((((..(......)..)))))))....(((......))).. ( -34.60) >consensus GAUUGGACUAGGGGUCCCCACCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUGGGAGUCAGACUGCGAGUGAUAAGAUCCGUAGUCAAAAGGGAAACAGCCCAG (((((((...(((((((.....))))).))...)))))))................((((((........))))))(((((((..(......)..)))))))....(((......))).. (-34.39 = -34.12 + -0.28)

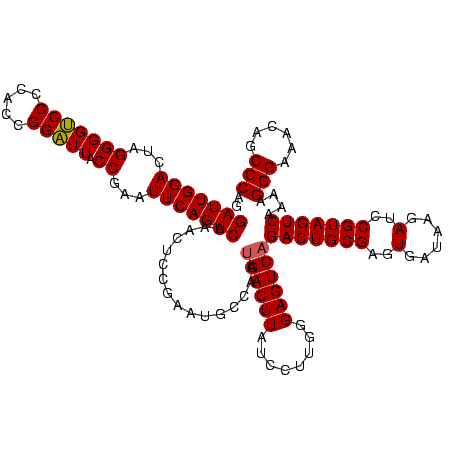
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -37.82 |
| Consensus MFE | -36.96 |
| Energy contribution | -36.52 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/1080-1200 GGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAU ..(((((((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..)))))((.(((.........))).))...)))).. ( -39.20) >Bsubt.0 947750 120 + 4214630/1080-1200 GGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAU ..(((((((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..)))))((.(((.........))).))...)))).. ( -39.20) >Bcere.0 606627 120 + 5224283/1080-1200 GGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAUACAC ...((((((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..)))))((.(((.........))).)).....))). ( -37.40) >Banth.0 534125 120 + 5227293/1080-1200 GGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAUACAC ...((((((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..)))))((.(((.........))).)).....))). ( -37.40) >Bhalo.0 119923 120 + 4202352/1080-1200 GGAUGUGGAGUUGCCCAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAU ..(((((((((..(...((...((((((((.(((((.......))))).))))....((((........)))).))))))..)..)))))((.(((.........))).))...)))).. ( -36.50) >Bclau.0 935920 120 + 4303871/1080-1200 GGAUGUGGAGUUGCCCAGACAACCAGGAUGUUGGCUUAGAAGCAGCCAUCAUUGAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAU ..(((((((((..(...((...((((((((.(((((.......))))).))))....((((........)))).))))))..)..)))))((.(((.........))).))...)))).. ( -37.20) >consensus GGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAU ..(((((((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..)))))((.(((.........))).))...)))).. (-36.96 = -36.52 + -0.44)

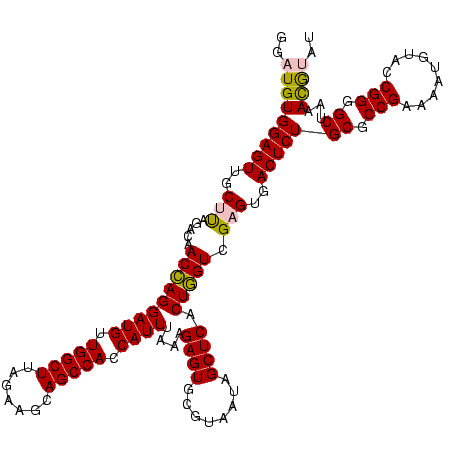
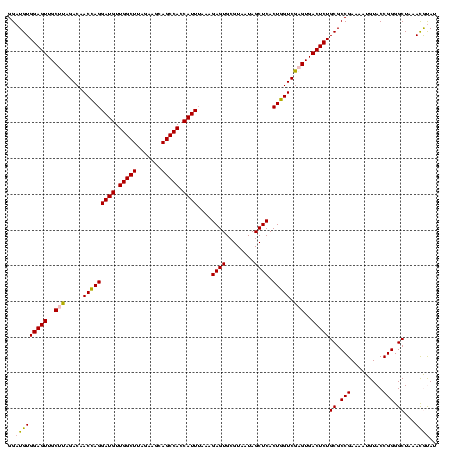
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.89 |
| Mean single sequence MFE | -32.58 |
| Consensus MFE | -30.90 |
| Energy contribution | -30.23 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980451 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/1080-1200 AUACGUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUAAGCAACUCCACAUCC ...((((((((.((((.......)))).))(((((((((.....)))))((......)).)))).))))))((((.(((((...(((((......)))))....)))))...)))).... ( -33.90) >Bsubt.0 947750 120 + 4214630/1080-1200 AUACGUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUAAGCAACUCCACAUCC ...((((((((.((((.......)))).))(((((((((.....)))))((......)).)))).))))))((((.(((((...(((((......)))))....)))))...)))).... ( -33.90) >Bcere.0 606627 120 + 5224283/1080-1200 GUGUAUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUAAGCAACUCCACAUCC (((((..((((.((((.......))))(((..((((....)))).))).)))).)))))............((((.(((((...((.((((......)))).)))))))...)))).... ( -31.30) >Banth.0 534125 120 + 5227293/1080-1200 GUGUAUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUAGUUGUCUAAGCAACUCCACAUCC (((((..((((.((((.......))))(((..((((....)))).))).)))).)))))............((((.(((((...((.((((......)))).)))))))...)))).... ( -31.30) >Bhalo.0 119923 120 + 4202352/1080-1200 AUACGUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUGGGCAACUCCACAUCC ...((((((((.((((.......)))).))(((((((((.....)))))((......)).)))).))))))((((.(((((...(((((......)))))....)))))...)))).... ( -33.50) >Bclau.0 935920 120 + 4303871/1080-1200 AUACGUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUCAAUGAUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUGGGCAACUCCACAUCC .........((.((((.......)))).))...(((...(((((((.((((......))..........((.(((((.......))))).)))))))))))....)))............ ( -31.60) >consensus AUACGUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUAAGCCAACAUCCUGGUUGUCUAAGCAACUCCACAUCC .........((.((((.......)))).)).((((..(((((((((.((((......))..........((.(((((.......))))).)))))))))))....))..))))....... (-30.90 = -30.23 + -0.66)

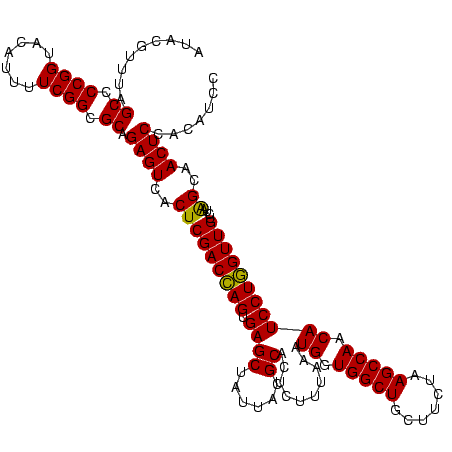
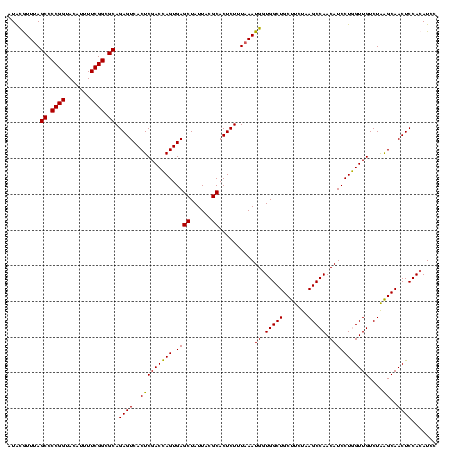
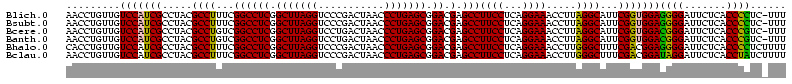
| Location | 922,209 – 922,326 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -45.43 |
| Consensus MFE | -44.31 |
| Energy contribution | -41.33 |
| Covariance contribution | -2.98 |
| Combinations/Pair | 1.49 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 117 + 4222334/1200-1320 CACCGAAGCUGCGGACUGUUCU---UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG .......(((((..(((((((.---...)))))))((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))).......))))). ( -48.70) >Bsubt.0 947750 116 + 4214630/1200-1320 CACCGAAGCUGCGGACUGUUCU---U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG .......(((((..(((((((.---.-.)))))))((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))).......))))). ( -47.80) >Bcere.0 606627 120 + 5224283/1200-1320 CACCGAAGCUGCGGAUUGAUACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG .......(((((..(((((((((...)))))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))).......))))). ( -41.00) >Banth.0 534125 120 + 5227293/1200-1320 CACCGAAGCUGCGGAUUGAUACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG .......(((((..(((((((((...)))))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))).......))))). ( -41.00) >Bhalo.0 119923 120 + 4202352/1200-1320 CACCGAAGCUAUGGAUUCGCACCGUUGGUGCGAGUGGUAGGGGAGCGUUCUACGUGCAGCGAAGUCAGACCGAGAGGACUGGUGGAGCGCGUAGAAGUGAGAAUGCCGGUAUGAGUAGCG .......(((((..(((((((((...)))))))))((((.....((.((((((((((..((...((((.((....)).))))))..)))))))))))).....)))).......))))). ( -47.90) >Bclau.0 935920 117 + 4303871/1200-1320 CACCGAAGCUAUGG--CAAUCCCGUAAGGGAUUG-GGUAGGGGAGCGUUCCAAGGACAGCGAAGCUAGAUCGUGAGGACUAGUGGAGUGCUUGGAAGUGAGAAUGCCGGUAUGAGUAGCG .......(((((..--(((((((....)))))))-((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))).......))))). ( -46.20) >consensus CACCGAAGCUGCGGAUUGAUACC__UGGGAACAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG .......(((((..((((((((.....))))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))).......))))). (-44.31 = -41.33 + -2.98)

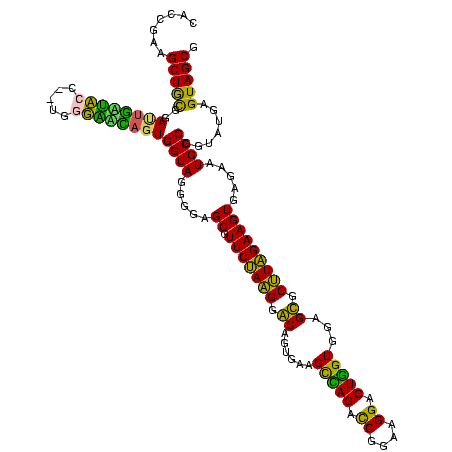
| Location | 922,209 – 922,326 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.53 |
| Mean single sequence MFE | -29.28 |
| Consensus MFE | -25.10 |
| Energy contribution | -22.89 |
| Covariance contribution | -2.22 |
| Combinations/Pair | 1.41 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.893900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 117 + 4222334/1200-1320 CGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCGCA---AGAACAGUCCGCAGCUUCGGUG .........((((((.........(((((((.(((...((((.((....)).))))......))).))))))).(((........(((((((...---.)))))))....))).)))))) ( -35.10) >Bsubt.0 947750 116 + 4214630/1200-1320 CGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCG-A---AGAACAGUCCGCAGCUUCGGUG .........((((((.........(((((((.(((...((((.((....)).))))......))).))))))).(((........(((((((.-.---.)))))))....))).)))))) ( -30.40) >Bcere.0 606627 120 + 5224283/1200-1320 CGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGGUAUCAAUCCGCAGCUUCGGUG .........(((((((........(((((((.((.....(((.((....)).)))........)).)))))))..............(((((((...))))))).......))..))))) ( -25.02) >Banth.0 534125 120 + 5227293/1200-1320 CGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGGUAUCAAUCCGCAGCUUCGGUG .........(((((((........(((((((.((.....(((.((....)).)))........)).)))))))..............(((((((...))))))).......))..))))) ( -25.02) >Bhalo.0 119923 120 + 4202352/1200-1320 CGCUACUCAUACCGGCAUUCUCACUUCUACGCGCUCCACCAGUCCUCUCGGUCUGACUUCGCUGCACGUAGAACGCUCCCCUACCACUCGCACCAACGGUGCGAAUCCAUAGCUUCGGUG .........(((((((........(((((((.((.....(((.((....)).)))........)).)))))))..............(((((((...))))))).......))..))))) ( -32.72) >Bclau.0 935920 117 + 4303871/1200-1320 CGCUACUCAUACCGGCAUUCUCACUUCCAAGCACUCCACUAGUCCUCACGAUCUAGCUUCGCUGUCCUUGGAACGCUCCCCUACC-CAAUCCCUUACGGGAUUG--CCAUAGCUUCGGUG .........((((((.........(((((((.((....((((((.....)).)))).......)).))))))).(((........-(((((((....)))))))--....))).)))))) ( -27.40) >consensus CGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGCCCUUAGAACGCUCCCCUACCACUGUUACCA__GGGAUCAAUCCGCAGCUUCGGUG .........(((((((........(((((((.((.....(((.((....)).)))........)).)))))))..............((((((.....)))))).......))..))))) (-25.10 = -22.89 + -2.22)
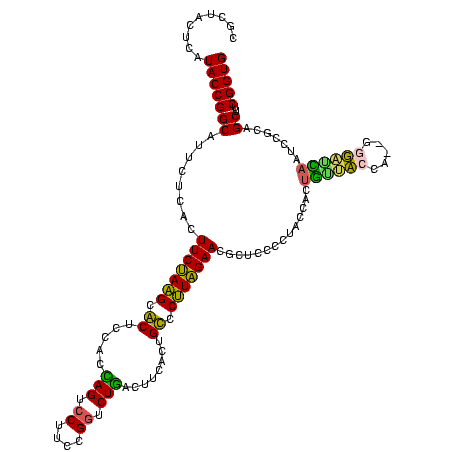
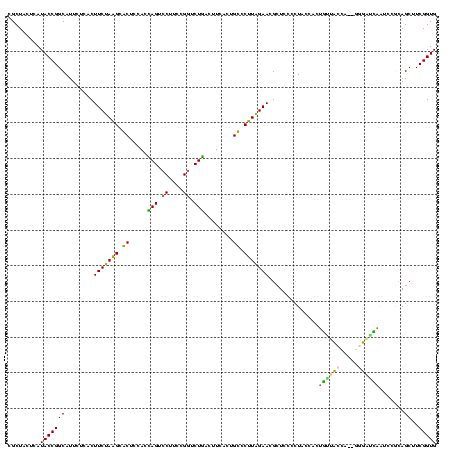
| Location | 922,209 – 922,328 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.52 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -37.53 |
| Energy contribution | -34.32 |
| Covariance contribution | -3.21 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 119 + 4222334/1240-1360 GAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAA-GAGGGGUGAGAAUCCCCUCCACCGAAUGCCUAAGGU ....((.(((((((.(((.(...(((((.((....)).)))))).))).)))))))))......((((((((......((...-((((((.......))))))...)).)))))...))) ( -43.30) >Bsubt.0 947750 119 + 4214630/1240-1360 GAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAA-GAGGGGUGAGAAUCCCCUCCACCGAAUGCCUAAGGU ....((.(((((((.(((.(...(((((.((....)).)))))).))).)))))))))......((((((((......((...-((((((.......))))))...)).)))))...))) ( -43.90) >Bcere.0 606627 119 + 5224283/1240-1360 GGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAA-GACGGGUGAGAAUCCCGUCCACCGAAUGCCUAAGGU .((.((((((...((((......(((...((....))(((((((...((((....))))....))))))).............-)))(((.......)))))))...))))))))..... ( -36.40) >Banth.0 534125 119 + 5227293/1240-1360 GGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAA-GACGGGUGAGAAUCCCGUCCACCGAAUGCCUAAGGU .((.((((((...((((......(((...((....))(((((((...((((....))))....))))))).............-)))(((.......)))))))...))))))))..... ( -36.40) >Bhalo.0 119923 120 + 4202352/1240-1360 GGGAGCGUUCUACGUGCAGCGAAGUCAGACCGAGAGGACUGGUGGAGCGCGUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAAGAGGGGUGAGAAUCCCCUCCGUCGAAAGCCCAAGGU (((.((.((((((((((..((...((((.((....)).))))))..))))))))))))....................(((...((((((.......))))))..)))....)))..... ( -42.10) >Bclau.0 935920 120 + 4303871/1240-1360 GGGAGCGUUCCAAGGACAGCGAAGCUAGAUCGUGAGGACUAGUGGAGUGCUUGGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAAGAUAGGUGAGAAUCCUAUCCGUCGAAAGCCCAAGGU (((.((((((((....(((((..(((((.((....)).)))))....))).))....)).))))))............(((...((((((.......))))))..)))....)))..... ( -37.30) >consensus GGGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAA_GAGGGGUGAGAAUCCCCUCCACCGAAUGCCUAAGGU .((.((.(((((((.((......(((((.((....)).)))))...)).)))))))(((.......((.((....)).))....((((((.......))))))))).....))))..... (-37.53 = -34.32 + -3.21)

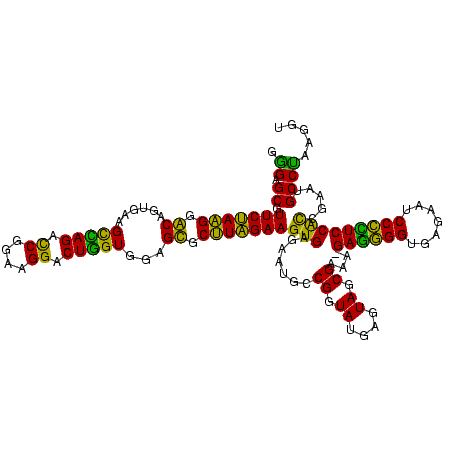
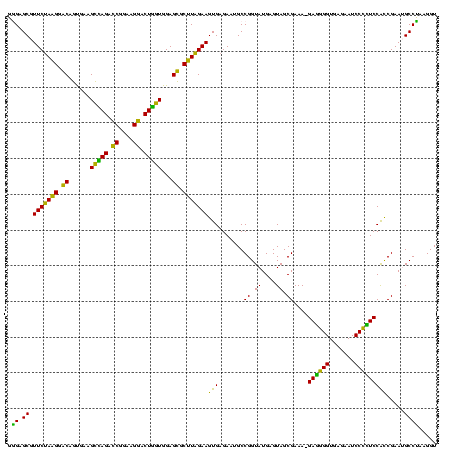
| Location | 922,209 – 922,328 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.52 |
| Mean single sequence MFE | -31.03 |
| Consensus MFE | -27.75 |
| Energy contribution | -25.25 |
| Covariance contribution | -2.49 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.979565 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 119 + 4222334/1240-1360 ACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUC-UUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUC ......(((..((((((((((((.......))))))-)............))))).........(((((((.(((...((((.((....)).))))......))).))))))).)))... ( -34.90) >Bsubt.0 947750 119 + 4214630/1240-1360 ACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUC-UUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUC ......(((..((((((((((((.......))))))-)............))))).........(((((((.(((...((((.((....)).))))......))).))))))).)))... ( -35.50) >Bcere.0 606627 119 + 5224283/1240-1360 ACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUC-UUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCC ......(((..((((((((((((.......))))))-)............))))).........(((((((.((.....(((.((....)).)))........)).))))))).)))... ( -28.52) >Banth.0 534125 119 + 5227293/1240-1360 ACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUC-UUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCC ......(((..((((((((((((.......))))))-)............))))).........(((((((.((.....(((.((....)).)))........)).))))))).)))... ( -28.52) >Bhalo.0 119923 120 + 4202352/1240-1360 ACCUUGGGCUUUCGACGGAGGGGAUUCUCACCCCUCUUUUCGCUACUCAUACCGGCAUUCUCACUUCUACGCGCUCCACCAGUCCUCUCGGUCUGACUUCGCUGCACGUAGAACGCUCCC .....((((....((.(((((((.......)))))))..))(((.........)))........(((((((.((.....(((.((....)).)))........)).))))))).)))).. ( -33.72) >Bclau.0 935920 120 + 4303871/1240-1360 ACCUUGGGCUUUCGACGGAUAGGAUUCUCACCUAUCUUUUCGCUACUCAUACCGGCAUUCUCACUUCCAAGCACUCCACUAGUCCUCACGAUCUAGCUUCGCUGUCCUUGGAACGCUCCC .....((((....((.(((((((.......)))))))..))(((.........)))........(((((((.((....((((((.....)).)))).......)).))))))).)))).. ( -25.00) >consensus ACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUC_UUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGCCCUUAGAACGCUCCC ......(((....((..((((((.......))))))...))(((.........)))........(((((((.((.....(((.((....)).)))........)).))))))).)))... (-27.75 = -25.25 + -2.49)

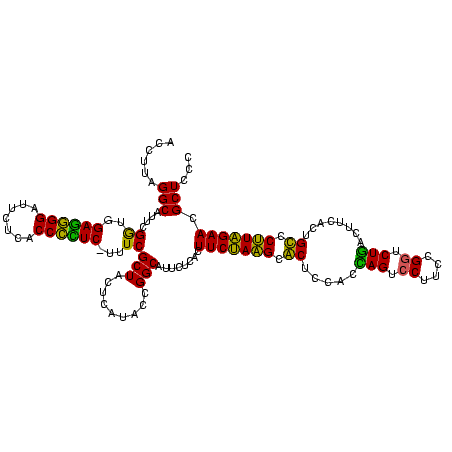
| Location | 922,209 – 922,328 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -44.92 |
| Consensus MFE | -44.79 |
| Energy contribution | -43.90 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.60 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.998630 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 119 + 4222334/1320-1440 AAA-GAGGGGUGAGAAUCCCCUCCACCGAAUGCCUAAGGUUUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUU ...-((((((.......))))))........((((.((.(((((...))))).)).(((((.((.(((((......)))))..(((.(.(((....))).).))))))))))...)))). ( -47.20) >Bsubt.0 947750 119 + 4214630/1320-1440 AAA-GAGGGGUGAGAAUCCCCUCCACCGAAUGCCUAAGGUUUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUU ...-((((((.......))))))........((((.((.(((((...))))).)).(((((.((.(((((......)))))..(((.(.(((....))).).))))))))))...)))). ( -47.20) >Bcere.0 606627 119 + 5224283/1320-1440 AAA-GACGGGUGAGAAUCCCGUCCACCGAAUGCCUAAGGUUUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCAGGACCUAAGCCGAGGCCGACAGGCGUAGGCGAUGGACAACAGGUU ...-((((((.......))))))........((((.((.(((((...))))).)).(((((.((.(((((......)))))..(((.(.(((....))).).))))))))))...)))). ( -43.40) >Banth.0 534125 119 + 5227293/1320-1440 AAA-GACGGGUGAGAAUCCCGUCCACCGAAUGCCUAAGGUUUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCAGGACCUAAGCCGAGGCCGACAGGCGUAGGCGAUGGACAACAGGUU ...-((((((.......))))))........((((.((.(((((...))))).)).(((((.((.(((((......)))))..(((.(.(((....))).).))))))))))...)))). ( -43.40) >Bhalo.0 119923 120 + 4202352/1320-1440 AAAAGAGGGGUGAGAAUCCCCUCCGUCGAAAGCCCAAGGUUUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUG ....((((((.......))))))........(((..((.(((((...))))).)).(((((.((.(((((......)))))..(((.(.(((....))).).))))))))))....))). ( -46.30) >Bclau.0 935920 120 + 4303871/1320-1440 AAAAGAUAGGUGAGAAUCCUAUCCGUCGAAAGCCCAAGGUUUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUU ....((((((.......)))))).......((((..((.(((((...))))).)).(((((.((.(((((......)))))..(((.(.(((....))).).))))))))))....)))) ( -42.00) >consensus AAA_GAGGGGUGAGAAUCCCCUCCACCGAAUGCCUAAGGUUUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUU ....((((((.......))))))........(((..((.(((((...))))).)).(((((.((.(((((......)))))..(((.(.(((....))).).))))))))))....))). (-44.79 = -43.90 + -0.89)

| Location | 922,209 – 922,328 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.82 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -38.60 |
| Energy contribution | -37.60 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 119 + 4222334/1320-1440 AACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUC-UUU .(((..((((((.........(((....)))....(((((((...........)))))))))))))((((......(....)...))))....)))(((((((.......))))))-).. ( -42.40) >Bsubt.0 947750 119 + 4214630/1320-1440 AACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUC-UUU .(((..((((((.........(((....)))....(((((((...........)))))))))))))((((......(....)...))))....)))(((((((.......))))))-).. ( -42.40) >Bcere.0 606627 119 + 5224283/1320-1440 AACCUGUUGUCCAUCGCCUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUC-UUU .(((..((((((...(((.((....)).)))....(((((((...........)))))))))))))((((......(....)...))))....)))(((((((.......))))))-).. ( -42.40) >Banth.0 534125 119 + 5227293/1320-1440 AACCUGUUGUCCAUCGCCUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUC-UUU .(((..((((((...(((.((....)).)))....(((((((...........)))))))))))))((((......(....)...))))....)))(((((((.......))))))-).. ( -42.40) >Bhalo.0 119923 120 + 4202352/1320-1440 CACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUGGGCUUUCGACGGAGGGGAUUCUCACCCCUCUUUU .....(((((((.........(((....)))....(((((((...........)))))))))))((((((......(....)...))))))..)))(((((((.......)))))))... ( -40.60) >Bclau.0 935920 120 + 4303871/1320-1440 AACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUGGGCUUUCGACGGAUAGGAUUCUCACCUAUCUUUU .....(((((((.........(((....)))....(((((((...........)))))))))))((((((......(....)...))))))..)))(((((((.......)))))))... ( -36.40) >consensus AACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUC_UUU .........(((((((.....((((...((((((.(((((((...........))))))).)).).)))((((...)))).....))))...)))))))((((.......))))...... (-38.60 = -37.60 + -1.00)
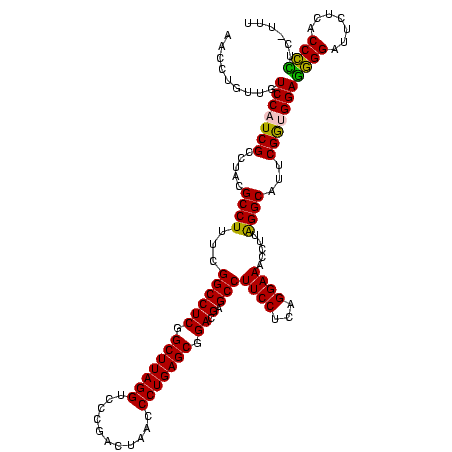
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -44.92 |
| Consensus MFE | -42.58 |
| Energy contribution | -42.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964427 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/1360-1480 UUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUUGAUAUUCCUGUACCACCUCCUCACCAUUUGAGCAAUGGGG ....((((((.((...(((((.((.(((((......)))))..(((.(.(((....))).).)))))))))).(((((........)))))....))))))))(((((.....))))).. ( -48.20) >Bsubt.0 947750 120 + 4214630/1360-1480 UUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUUGAUAUUCCUGUACCACCUCCUCACCAUUUGAGCAAUGGGG ....((((((.((...(((((.((.(((((......)))))..(((.(.(((....))).).)))))))))).(((((........)))))....))))))))(((((.....))))).. ( -48.20) >Bcere.0 606627 120 + 5224283/1360-1480 UUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCAGGACCUAAGCCGAGGCCGACAGGCGUAGGCGAUGGACAACAGGUUGAUAUUCCUGUACCACCUCUUUAUCGUUUGAGCAAUGGAG .(((.((((..((...(((((.((.(((((......)))))..(((.(.(((....))).).)))))))))).(((((........))))).)).))))......((....))...))). ( -39.80) >Banth.0 534125 120 + 5227293/1360-1480 UUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCAGGACCUAAGCCGAGGCCGACAGGCGUAGGCGAUGGACAACAGGUUGAUAUUCCUGUACCACCUCUUUAUCGUUUGAGCAAUGGAG .(((.((((..((...(((((.((.(((((......)))))..(((.(.(((....))).).)))))))))).(((((........))))).)).))))......((....))...))). ( -39.80) >Bhalo.0 119923 120 + 4202352/1360-1480 UUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUGGAAAUUCCUGUACCACCUCCUCACCGUUUGAGCAAUGGGG ....((((((.((...(((((.((.(((((......)))))..(((.(.(((....))).).)))))))))).(((((........)))))....))))))))(((((.....))))).. ( -47.80) >Bclau.0 935920 120 + 4303871/1360-1480 UUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUUGAUAUUCCUGUACCACCUCGUAUUCGUUUGAACGAUGGGG ..(((((((..((((..(((.....)))..))))((..((...(((.(.(((....))).).)))...))...(((((........))))).)).))))....((((....)))).))). ( -45.70) >consensus UUCCUGAGGAAGGCUCGUCCGCUCAGGGUUAGUCGGGACCUAAGCCGAGGCCGAAAGGCGUAGGCGAUGGACAACAGGUUGAUAUUCCUGUACCACCUCCUCACCGUUUGAGCAAUGGGG ....((((((.((...(((((.((.(((((......)))))..(((.(.(((....))).).)))))))))).(((((........)))))....))))))))(((((.....))))).. (-42.58 = -42.13 + -0.44)
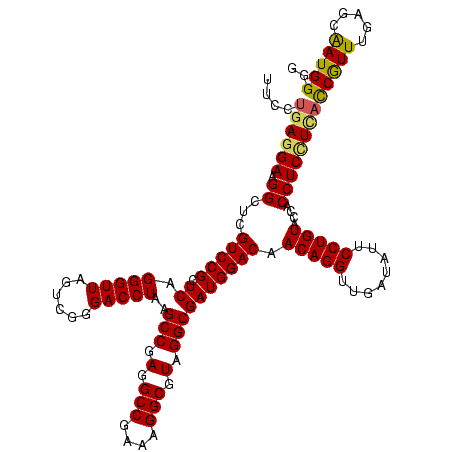
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.11 |
| Mean single sequence MFE | -42.07 |
| Consensus MFE | -36.98 |
| Energy contribution | -36.53 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.935465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/1360-1480 CCCCAUUGCUCAAAUGGUGAGGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAA (((((((.....)))))(((((((((((.(((((........)))))...))))).............((((((.(((((((...........))))))).)).).))).)))))))).. ( -44.40) >Bsubt.0 947750 120 + 4214630/1360-1480 CCCCAUUGCUCAAAUGGUGAGGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAA (((((((.....)))))(((((((((((.(((((........)))))...))))).............((((((.(((((((...........))))))).)).).))).)))))))).. ( -44.40) >Bcere.0 606627 120 + 5224283/1360-1480 CUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAA .(((((((......))))...((((.(((.((((........))))((((((...(((.((....)).)))....(((((((...........))))))))))))))))..)))).))). ( -39.90) >Banth.0 534125 120 + 5227293/1360-1480 CUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAA .(((((((......))))...((((.(((.((((........))))((((((...(((.((....)).)))....(((((((...........))))))))))))))))..)))).))). ( -39.90) >Bhalo.0 119923 120 + 4202352/1360-1480 CCCCAUUGCUCAAACGGUGAGGAGGUGGUACAGGAAUUUCCACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAA ..((...........(((((((.(((((.(((((........)))))...))))).))).))))....((((((.(((((((...........))))))).)).).))).......)).. ( -43.70) >Bclau.0 935920 120 + 4303871/1360-1480 CCCCAUCGUUCAAACGAAUACGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAA ..((.((((....))))....((((.(((.((((........))))((((((.........(((....)))....(((((((...........))))))))))))))))..)))).)).. ( -40.10) >consensus CCCCAUUGCUCAAACGAUGAGGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAA ..((((((......))))...((((.(((.((((........))))((((((.........(((....)))....(((((((...........))))))))))))))))..)))).)).. (-36.98 = -36.53 + -0.44)

| Location | 922,209 – 922,326 |
|---|---|
| Length | 117 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -29.39 |
| Energy contribution | -26.23 |
| Covariance contribution | -3.16 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
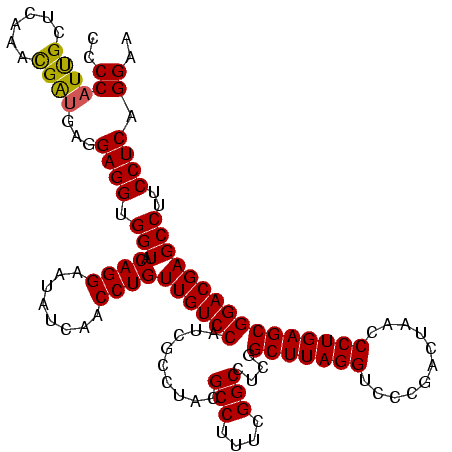
>Blich.0 922209 117 + 4222334/1480-1600 GGACGCAGGAGGAUAGGGCAAGCGCGGUAUUGGAUAUCCGCGUCCAAGCA-GUUAGGCUGGGAAAUAGGCAAAUCCGUUUCCCGU-AAGGCUGAGCUGUGAUGGCGA-GCGAAAUAUAGU ((((((.(((.((((..((....))..)))).....)))))))))..(((-(((.(((((((((((.((.....)))))))))..-..)))).))))))........-............ ( -37.40) >Bsubt.0 947750 117 + 4214630/1480-1600 GGACGCAGGAGGAUAGGGUAAGCGCGGUAUUGGAUAUCCGCGUCCAAGCA-GUUAGGCUGGGAAAUAGGCAAAUCCGUUUCCCAU-AAGGCUGAGCUGUGAUGGCGA-GCGAAAUAUAGU ...(((...............((((((..........))))))(((.(((-(((.(((((((((((.((.....)))))))))..-..)))).))))))..)))...-)))......... ( -36.60) >Bcere.0 606627 118 + 5224283/1480-1600 GGACGCAGAAGGAUAGAAGAAGCGUGCGAUUGGUUGUGCACGUCCAAGCA-GUUAGGCUGAUAAGUAGGCAAAUCCGCUUAUCGUAAAGGCUGAGCUGUGAUGGGGAAGCUCCUUAUGG- .....((.(((((........(((((((........)))))))(((.(((-(((.(((((((((((.((.....))))))))).....)))).))))))..)))......))))).)).- ( -36.40) >Banth.0 534125 118 + 5227293/1480-1600 GGCCGCAGAAGGAUAGAAGAAGCGUGCGAUUGGUUGUGCACGUCCAAGCA-GUUAGGCUGAUAAGUAGGCAAAUCCGCUUAUCGUGAAGGCUGAGCUGUGAUGGGGAAGCUCCUUAUGG- .....((.(((((........(((((((........)))))))(((.(((-(((.(((((((((((.((.....))))))))).....)))).))))))..)))......))))).)).- ( -36.40) >Bhalo.0 119923 119 + 4202352/1480-1600 GGACGCAGAAAGGUAGGGUAAGCGCGCUGAUGGAGAUGCGCGUCCAAGCA-GUUAGGCUGGAAAGUAGGCAAAUCCGCUUUCCAUAAAGGCUGAGCUGUGAUGGCGAGGGAAAUAAUAGU ...(((..........((...(((((((.....)).)))))..))..(((-(((.(((((((((((.((.....))))))))).....)))).))))))....))).............. ( -35.10) >Bclau.0 935920 119 + 4303871/1480-1600 GGACGCAGAAAGGUAGGGUGAGCGCGCCGCUGGCUGUGCGCGUCGAAGCAUGCGAGGCUGGAACAUAGGCAAAUCCGUGUUCCAUAA-GGCUGAGCUGUGACCGUGAAGGACCUUCGGGU ((.(((((..(((((.(((.((((...)))).))).)))((.(((.......))).))((((((((.((.....))))))))))...-..))...))))).)).......(((....))) ( -44.00) >consensus GGACGCAGAAGGAUAGGGUAAGCGCGCUAUUGGAUAUGCGCGUCCAAGCA_GUUAGGCUGGAAAAUAGGCAAAUCCGCUUACCAUAAAGGCUGAGCUGUGAUGGCGAAGCAAAUUAUAGU ...(((((..((.........((((((.((.....))))))))((.(((.......)))(((((((.((.....))))))))).....))))...))))).................... (-29.39 = -26.23 + -3.16)
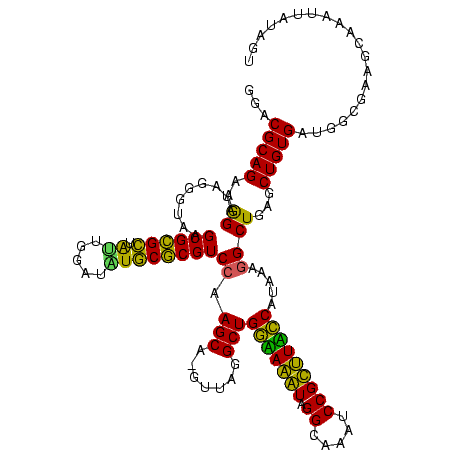
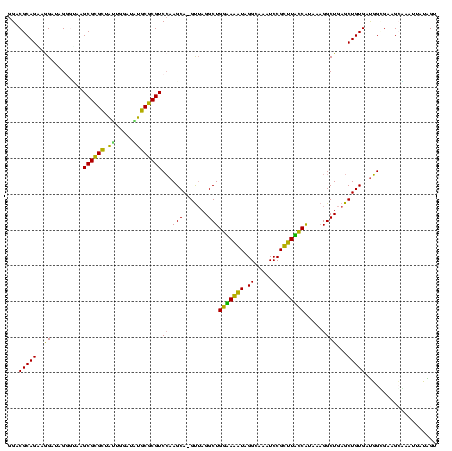
| Location | 922,209 – 922,325 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.09 |
| Mean single sequence MFE | -45.95 |
| Consensus MFE | -38.40 |
| Energy contribution | -37.93 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929077 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 116 + 4222334/1840-1960 UGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGC----GUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAA ((((..(((...(((((.(((......)))..)))))(((...((((((..((((..((...((((((.((----....))..))))))...))..))))))))))...)))))))))). ( -47.90) >Bsubt.0 947750 116 + 4214630/1840-1960 UGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGC----GUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAA ((((..(((...(((((.(((......)))..)))))(((...((((((..((((..((...((((((.((----....))..))))))...))..))))))))))...)))))))))). ( -47.90) >Bcere.0 606627 116 + 5224283/1840-1960 UGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC----GUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAA (((...(((....)))...)))((.(((....)..(((((...((((((..((((..((...((((((.((----....))..))))))...))..))))))))))...))))))).)). ( -45.40) >Banth.0 534125 116 + 5227293/1840-1960 UGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC----GUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAA (((...(((....)))...)))((.(((....)..(((((...((((((..((((..((...((((((.((----....))..))))))...))..))))))))))...))))))).)). ( -45.40) >Bhalo.0 119923 119 + 4202352/1840-1960 UGCGAAGCCGCAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAGGGGUUAUCCCUU-CGGGAGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAA ((((..(((((..............((((........))))..((((((..((((..((...((((((.((((..-..)).))))))))...))..))))))))))....))))))))). ( -44.90) >Bclau.0 935920 120 + 4303871/1840-1960 UGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAAGGGUCAUCCCUUACGGGAGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAA ((((..(((((..............((((........))))..((((((..((((..((...(((((..((((....))))...)))))...))..)))))))))).)))))...)))). ( -44.20) >consensus UGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAGGGGUUAGC____GUAAGCGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAA ((((..(((....)))...................(((((...((((((..((((..((...(((((..((........))...)))))...))..))))))))))...))))).)))). (-38.40 = -37.93 + -0.47)

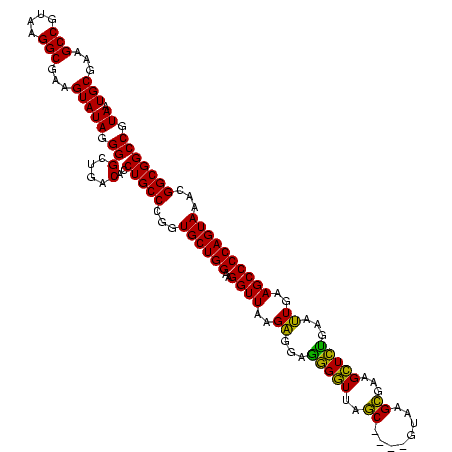
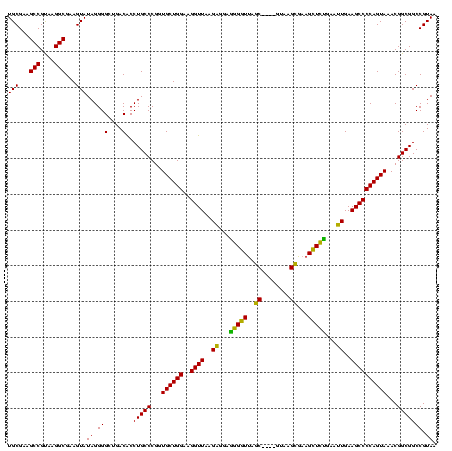
| Location | 922,209 – 922,325 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.27 |
| Mean single sequence MFE | -38.32 |
| Consensus MFE | -35.50 |
| Energy contribution | -33.37 |
| Covariance contribution | -2.13 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 116 + 4222334/1880-2000 CGGUGCUGGAAGGUUAAGAGGAGCGCUUAGC----GUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAA ...((((((..((((..((...((((((.((----....))..))))))...))..))))))))))..((((((((((.......))))))..((((..........))))))))..... ( -39.00) >Bsubt.0 947750 116 + 4214630/1880-2000 CGGUGCUGGAAGGUUAAGAGGAGCGCUUAGC----GUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAA ...((((((..((((..((...((((((.((----....))..))))))...))..))))))))))..((((((((((.......))))))..((((..........))))))))..... ( -39.00) >Bcere.0 606627 116 + 5224283/1880-2000 CGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC----GUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAA ...((((((..((((..((...((((((.((----....))..))))))...))..))))))))))..((((((((((.......))))))..((((..........))))))))..... ( -37.50) >Banth.0 534125 116 + 5227293/1880-2000 CGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC----GUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAA ...((((((..((((..((...((((((.((----....))..))))))...))..))))))))))..((((((((((.......))))))..((((..........))))))))..... ( -37.50) >Bhalo.0 119923 119 + 4202352/1880-2000 CGGUGCUGGAAGGUUAAGAGGAGGGGUUAUCCCUU-CGGGAGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAA ...((((((..((((..((...((((((.((((..-..)).))))))))...))..))))))))))..((((((((((.......))))))..((((..........))))))))..... ( -38.40) >Bclau.0 935920 120 + 4303871/1880-2000 CGGUGCUGGAAGGUUAAGAGGAAGGGUCAUCCCUUACGGGAGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAA ...((((((..((((..((...(((((..((((....))))...)))))...))..))))))))))..((((((((((.......))))))..((((..........))))))))..... ( -38.50) >consensus CGGUGCUGGAAGGUUAAGAGGAGGGGUUAGC____GUAAGCGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAA ...((((((..((((..((...(((((..((........))...)))))...))..))))))))))..((((((((((.......))))))..((((..........))))))))..... (-35.50 = -33.37 + -2.13)

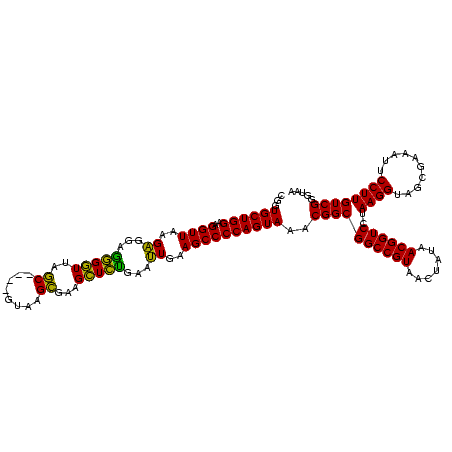
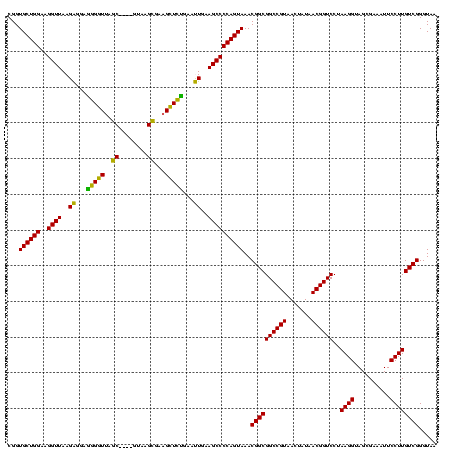
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -40.07 |
| Consensus MFE | -38.71 |
| Energy contribution | -38.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2000-2120 GCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAGAUCGUUGCGCCUUUCGUGCGGGUCGGAAC ((((....))))..(((((.((((.((((((.....((((...((...((((((....(((((.((((...)))).))))).))))))..))....))))...)))))))))).))))). ( -40.80) >Bsubt.0 947750 120 + 4214630/2000-2120 GCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAGAUCGUUGCGCCUUUCGUGCGGGUCGGAAC ((((....))))..(((((.((((.((((((.....((((...((...((((((....(((((.((((...)))).))))).))))))..))....))))...)))))))))).))))). ( -40.80) >Bcere.0 606627 120 + 5224283/2000-2120 GCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUGGUUGAGACAGUGCCCAAAUCGUUACACCUUUCGUGCGGGUCGGAAC ((((....))))..(((((.((((.(((((((((((((........)))))................(((((.....)))))..))))).....(.....).....))))))).))))). ( -39.10) >Banth.0 534125 120 + 5227293/2000-2120 GCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUGGUUGAGACAGUGCCCAAAUCGUUACACCUUUCGUGCGGGUCGGAAC ((((....))))..(((((.((((.(((((((((((((........)))))................(((((.....)))))..))))).....(.....).....))))))).))))). ( -39.10) >Bhalo.0 119923 120 + 4202352/2000-2120 GCUCCAUGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGGGUCUCUCGUUGAGACAGUAUCCAAGUCGUUACACCAUUCGUGCGGGUCGGAAC ((((....))))..(((((.((((.(((((((.(((((........)))))..............(((((((.....))))).(((..........))).))))))))))))).))))). ( -40.30) >Bclau.0 935920 120 + 4303871/2000-2120 GCUCCAUGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGGGUCUCUCGUUGAGACAGUAUCCAAGUCGUUGCACCAUUCGUGCGGGUCGGAAC ((((....))))..(((((.((((.(((((((.(((((........)))))..............(((((((.....))))).(((..........))).))))))))))))).))))). ( -40.30) >consensus GCUCCACGGGGUCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCGUUACACCUUUCGUGCGGGUCGGAAC ((((....))))..(((((.((((.(((((((((((((........)))))................(((((.....)))))..))))).....(.....).....))))))).))))). (-38.71 = -38.27 + -0.44)
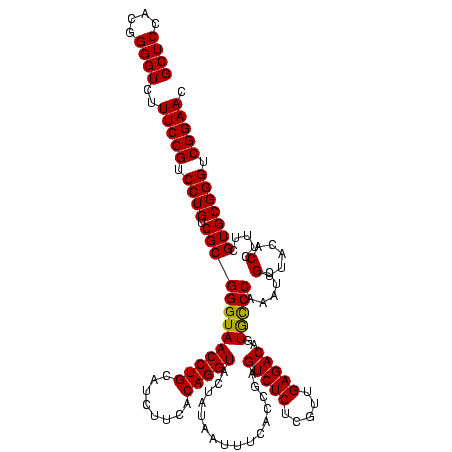
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.83 |
| Mean single sequence MFE | -46.38 |
| Consensus MFE | -46.91 |
| Energy contribution | -43.53 |
| Covariance contribution | -3.38 |
| Combinations/Pair | 1.47 |
| Mean z-score | -2.84 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.67 |
| SVM RNA-class probability | 0.999936 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2120-2240 UUUACUGCAGCCUGAUAUUGAAUGUUGGUACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACC .(((.(((((((.(((.(((..((((.((((.....))))..))))..)))..(((((....((((((((....)))))))).))))))))(((((....)))))..))))))).))).. ( -46.50) >Bsubt.0 947750 120 + 4214630/2120-2240 UUUACUGCAGCCUGAUAUUGAAUGUUGGUACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACC .(((.(((((((.(((.(((..((((.((((.....))))..))))..)))..(((((....((((((((....)))))))).))))))))(((((....)))))..))))))).))).. ( -46.50) >Bcere.0 606627 120 + 5224283/2120-2240 UUUACUGUAGCCUGAUAUUGAAUUUUGGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAA ...((((((.((.((........)).))))))))..((((((..(((((((..(((((....((((((((....)))))))).))))).)))((((....)))))))).))))))..... ( -45.00) >Banth.0 534125 120 + 5227293/2120-2240 UUUACUGUAGCCUGAUAUUGAAUUUUGGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAA ...((((((.((.((........)).))))))))..((((((..(((((((..(((((....((((((((....)))))))).))))).)))((((....)))))))).))))))..... ( -45.00) >Bhalo.0 119923 120 + 4202352/2120-2240 UUUACUGUAGCUUGAUAUUGGAUGUUGGUACCGUUUGUACAGGAUAGGUAGGAGCCUUGGAAGCCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACUACCCUGACGGUAUUGACA ......................(((..(((((((.....(((........((.(((((....((((((((....)))))))).))))).))(((((....)))))))))))))))..))) ( -45.00) >Bclau.0 935920 120 + 4303871/2120-2240 UUUACUGUAGCUUGAUAGUGAGUGUUGGUAUCAUCUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCUAGCUUCGGUGGAGGCGUCGGUGGGAUACUACCCUGAUGGUGCUGGCA ((((((((......))))))))(((..((((((((...............(..(((((....((((((((....)))))))).)))))..)(((((....)))))..))))))))..))) ( -50.30) >consensus UUUACUGUAGCCUGAUAUUGAAUGUUGGUACAGUUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCCGGUGGGAUACUACCCUGACUGUAUUGACA .......................((..((((((((...............((.(((((....((((((((....)))))))).))))).))(((((....)))))..))))))))..)). (-46.91 = -43.53 + -3.38)

| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -49.92 |
| Consensus MFE | -49.17 |
| Energy contribution | -45.77 |
| Covariance contribution | -3.41 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985338 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2160-2280 AGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCA ...((((.((((((((((....((((((((....)))))))).))))).))(((((....)))))))).))))..(((((((((...((.((((((.....))))))))))).)).)))) ( -53.10) >Bsubt.0 947750 120 + 4214630/2160-2280 AGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUGUCA ...((((.((((((((((....((((((((....)))))))).))))).))(((((....)))))))).))))..(((((((((...((.((((((.....))))))))))).)).)))) ( -54.60) >Bcere.0 606627 120 + 5224283/2160-2280 AGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCA ....((((..((((((((....((((((((....)))))))).)))))...(((((....)))))................)))..))))((((((.....))))))(....)....... ( -48.60) >Banth.0 534125 120 + 5227293/2160-2280 AGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCA ....((((..((((((((....((((((((....)))))))).)))))...(((((....)))))................)))..))))((((((.....))))))(....)....... ( -48.60) >Bhalo.0 119923 120 + 4202352/2160-2280 AGGAUAGGUAGGAGCCUUGGAAGCCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACUACCCUGACGGUAUUGACAUUCUAACCUCGAACCGUGAUCCGGUUCAGGGACAGUGUCA .............(((((....((((((((....)))))))).)))))((((..(((......)))...))))..(((((((....(((.((((((.....)))))))))...))))))) ( -48.30) >Bclau.0 935920 120 + 4303871/2160-2280 AGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCUAGCUUCGGUGGAGGCGUCGGUGGGAUACUACCCUGAUGGUGCUGGCAUUCUAACCUCGACCCGUGAUCCGGGCCAGGGACAUUGUCA ..(((((.((((((((((....((((((((....)))))))).)))))(((((((((......))).......)))))).))))).(((.(.((((.....))))).)))....))))). ( -46.31) >consensus AGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCCGGUGGGAUACUACCCUGACUGUAUUGACAUUCUAACCUACAGCCCUUAUCGGGCCGGGAGACAGUGUCA ...((((.(((..(((((....((((((((....)))))))).)))))...(((((....)))))))).))))..(((((((((...((.((((((.....)))))))))))..)))))) (-49.17 = -45.77 + -3.41)

| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.06 |
| Mean single sequence MFE | -39.57 |
| Consensus MFE | -38.33 |
| Energy contribution | -35.09 |
| Covariance contribution | -3.24 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.799452 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2160-2280 UGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCU ((((.(((.(((.((((((.....)))))).))).)))...))))....((((..((..(((((.....)))))..))...((((((((....))))))))...))))............ ( -44.00) >Bsubt.0 947750 120 + 4214630/2160-2280 UGACACUGUCUCCCCGCCCGAUAAGGGCGGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCU ((((.(((.(((.((((((.....)))))).))).)))...))))....((((..((..(((((.....)))))..))...((((((((....))))))))...))))............ ( -45.50) >Bcere.0 606627 120 + 5224283/2160-2280 UGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCU ((((.(((.((..((((((.....))))))..)).))).((....))...))))(((((((.....)))...((((..((((.(((....))).)))).....))))....))))..... ( -38.30) >Banth.0 534125 120 + 5227293/2160-2280 UGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCU ((((.(((.((..((((((.....))))))..)).))).((....))...))))(((((((.....)))...((((..((((.(((....))).)))).....))))....))))..... ( -38.30) >Bhalo.0 119923 120 + 4202352/2160-2280 UGACACUGUCCCUGAACCGGAUCACGGUUCGAGGUUAGAAUGUCAAUACCGUCAGGGUAGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGCUUCCAAGGCUCCUACCUAUCCU ((((((((.((.(((((((.....))))))).)).)))..))))).........((((((..........(.((((...(((.(((....))).)))......)))).)))))))..... ( -36.10) >Bclau.0 935920 120 + 4303871/2160-2280 UGACAAUGUCCCUGGCCCGGAUCACGGGUCGAGGUUAGAAUGCCAGCACCAUCAGGGUAGUAUCCCACCGACGCCUCCACCGAAGCUAGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCU (((...(((...(((((((.....))))))).(((......))).)))...)))((((((.........((.((((..((((.(((....))).)))).....))))))))))))..... ( -35.20) >consensus UGACACUGUCUCCCGGCCCGAUAAGGGCCGCAGGUUAGAAUGUCAAUACAGUCAGGGUAGUAUCCCACCAACGCCUCCACCGAAGCUAGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCU ((((.(((.((..((((((.....))))))..)).)))...)))).........((((((............((((..((((.(((....))).)))).....))))..))))))..... (-38.33 = -35.09 + -3.24)


| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.78 |
| Mean single sequence MFE | -49.00 |
| Consensus MFE | -47.49 |
| Energy contribution | -44.30 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.900752 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2200-2320 GUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUA ..((((((...(((((....)))))..((((((.((((((((((...((.((((((.....))))))))))).)).)))))....)))))).(((((...)))))))))))......... ( -49.90) >Bsubt.0 947750 120 + 4214630/2200-2320 GUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUA ..((((((...(((((....)))))..((((((.((((((((((...((.((((((.....))))))))))).)).)))))....)))))).(((((...)))))))))))......... ( -51.40) >Bcere.0 606627 120 + 5224283/2200-2320 GUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUA ..(((((((..........((((((((((((((.((((....((......((((((.....))))))(....)))..))))....)))))).....)))))))))))))))......... ( -49.30) >Banth.0 534125 120 + 5227293/2200-2320 GUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUA ..(((((((..........((((((((((((((.((((....((......((((((.....))))))(....)))..))))....)))))).....)))))))))))))))......... ( -49.30) >Bhalo.0 119923 120 + 4202352/2200-2320 GUGGAGGCGCUGGUGGGAUACUACCCUGACGGUAUUGACAUUCUAACCUCGAACCGUGAUCCGGUUCAGGGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAACAGUA ..((((((...(((((....)))))..(((.((.((((((((....(((.((((((.....)))))))))...)))))))).....(((.....)))..)).)))))))))......... ( -47.50) >Bclau.0 935920 120 + 4303871/2200-2320 GUGGAGGCGUCGGUGGGAUACUACCCUGAUGGUGCUGGCAUUCUAACCUCGACCCGUGAUCCGGGCCAGGGACAUUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAACAGUA ..((((((((((((((....)))))((((..((((((((..((.......)).(((.....))))))))...)))..))))...))).....(((((...)))))))))))......... ( -46.60) >consensus GUGGAGGCGCCGGUGGGAUACUACCCUGACUGUAUUGACAUUCUAACCUACAGCCCUUAUCGGGCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAAGUA ..((((((...(((((....)))))..((((((.((((((((((...((.((((((.....)))))))))))..))))))).....(((.....)))..))))))))))))......... (-47.49 = -44.30 + -3.19)

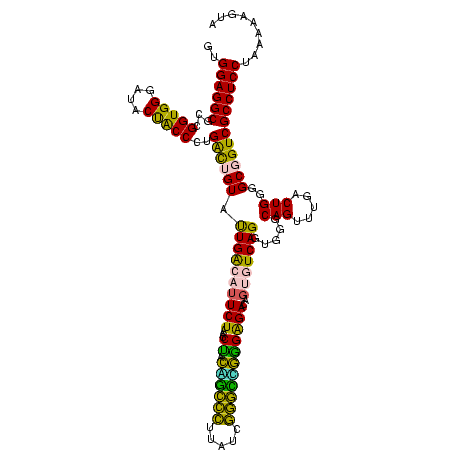
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 84.78 |
| Mean single sequence MFE | -45.57 |
| Consensus MFE | -44.65 |
| Energy contribution | -41.02 |
| Covariance contribution | -3.63 |
| Combinations/Pair | 1.43 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998725 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2200-2320 UACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCAC .........(((((((((.......)))..(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))...........)))))).. ( -47.20) >Bsubt.0 947750 120 + 4214630/2200-2320 UACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCCGCCCGAUAAGGGCGGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCAC .........(((((((((.......)))..(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))...........)))))).. ( -48.70) >Bcere.0 606627 120 + 5224283/2200-2320 UACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCAC .........((((((((((((((..((((...........)))).(((.((..((((((.....))))))..)).)))................)))))))..........))))))).. ( -45.70) >Banth.0 534125 120 + 5227293/2200-2320 UACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCAC .........((((((((((((((..((((...........)))).(((.((..((((((.....))))))..)).)))................)))))))..........))))))).. ( -45.70) >Bhalo.0 119923 120 + 4202352/2200-2320 UACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCCCUGAACCGGAUCACGGUUCGAGGUUAGAAUGUCAAUACCGUCAGGGUAGUAUCCCACCAGCGCCUCCAC .........(((((((((.......)))..(((((((((.((((((((.((.(((((((.....))))))).)).)))..))))).....))..)))))))...........)))))).. ( -43.70) >Bclau.0 935920 120 + 4303871/2200-2320 UACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACAAUGUCCCUGGCCCGGAUCACGGGUCGAGGUUAGAAUGCCAGCACCAUCAGGGUAGUAUCCCACCGACGCCUCCAC .........(((((((((.......)))..(((((((...(((...(((...(((((((.....))))))).(((......))).)))...))))))))))...........)))))).. ( -42.40) >consensus UACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGCCCGAUAAGGGCCGCAGGUUAGAAUGUCAAUACAGUCAGGGUAGUAUCCCACCAACGCCUCCAC .........(((((((((.......)))..((((((((((((((...)))...((((((.....)))))).))))..(.....)..........)))))))...........)))))).. (-44.65 = -41.02 + -3.63)
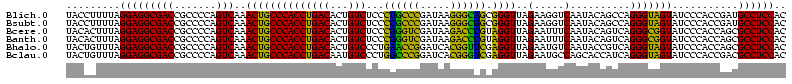
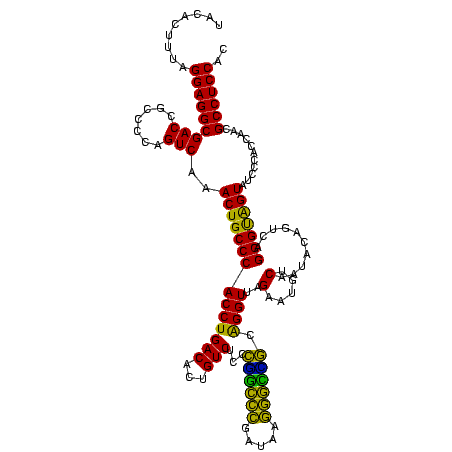

| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -50.23 |
| Consensus MFE | -49.68 |
| Energy contribution | -46.85 |
| Covariance contribution | -2.83 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.99 |
| SVM decision value | 4.12 |
| SVM RNA-class probability | 0.999804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2240-2360 UUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUC ((((((((.(((((((..(((.((((.(....)..)))).))))))))))....((((((.((.(((((((...........))))))).))........))))))...))))))))... ( -51.30) >Bsubt.0 947750 120 + 4214630/2240-2360 UUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUC ((((((((...(((((.....)))))(((((((...)))...(((((.....(((((...)))))((((((...........))))))))))).....)))).......))))))))... ( -51.40) >Bcere.0 606627 120 + 5224283/2240-2360 UUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUC ((((((((...(((((.....)))))(((((((...)))...(((((.....(((((...)))))((((((...........))))))))))).....)))).......))))))))... ( -51.30) >Banth.0 534125 120 + 5227293/2240-2360 UUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUC ((((((((...(((((.....)))))(((((((...)))...(((((.....(((((...)))))((((((...........))))))))))).....)))).......))))))))... ( -51.30) >Bhalo.0 119923 120 + 4202352/2240-2360 UUCUAACCUCGAACCGUGAUCCGGUUCAGGGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAACAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUC ((((((((..((((((.....))))))(((((......(...(((((.....(((((...)))))((((((...........)))))))))))...).)))))......))))))))... ( -49.00) >Bclau.0 935920 120 + 4303871/2240-2360 UUCUAACCUCGACCCGUGAUCCGGGCCAGGGACAUUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAACAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUC ((((((((..(.((((.....)))).)(((((.(((......(((((.....(((((...)))))((((((...........)))))))))))..))))))))......))))))))... ( -47.10) >consensus UUCUAACCUACAGCCCUUAUCGGGCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUC ((((((((..((((((.....)))))).(((.......(...(((((.....(((((...)))))((((((...........)))))))))))...)....))).....))))))))... (-49.68 = -46.85 + -2.83)

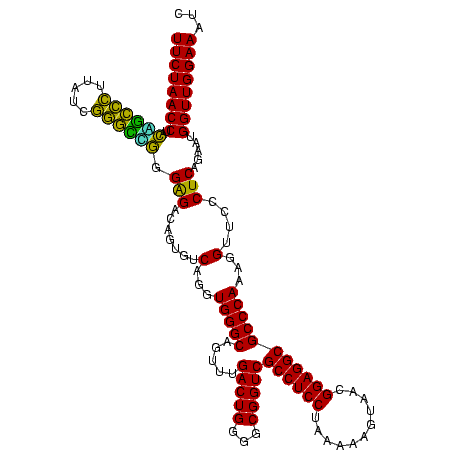
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -45.43 |
| Energy contribution | -42.24 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.48 |
| Structure conservation index | 1.01 |
| SVM decision value | 4.49 |
| SVM RNA-class probability | 0.999908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2240-2360 GAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAA ...(((.((((......(((((.....(((((((((((...........))))))(((.......))).....)))))...(((...))))))))((((.....))))....)))).))) ( -44.50) >Bsubt.0 947750 120 + 4214630/2240-2360 GAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCCGCCCGAUAAGGGCGGCGGGUUAGAA ...(((.((((.......((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....)))))...)))).))) ( -45.10) >Bcere.0 606627 120 + 5224283/2240-2360 GAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAA ...(((.((((.......((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....)))))...)))).))) ( -45.40) >Banth.0 534125 120 + 5227293/2240-2360 GAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAA ...(((.((((.......((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....)))))...)))).))) ( -45.40) >Bhalo.0 119923 120 + 4202352/2240-2360 GAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCCCUGAACCGGAUCACGGUUCGAGGUUAGAA ...(((.((((......(((((.....(((((((((((...........))))))(((.......))).....)))))..........)))))((((((.....))))))..)))).))) ( -44.26) >Bclau.0 935920 120 + 4303871/2240-2360 GAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACAAUGUCCCUGGCCCGGAUCACGGGUCGAGGUUAGAA ...(((.((((......(((((.....(((((((((((...........))))))(((.......))).....)))))..........)))))((((((.....))))))..)))).))) ( -46.06) >consensus GAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGCCCGAUAAGGGCCGCAGGUUAGAA ...(((.((((.......((((.....(((((((((((...........))))))(((.......))).....)))))..........)))).((((((.....))))))..)))).))) (-45.43 = -42.24 + -3.19)

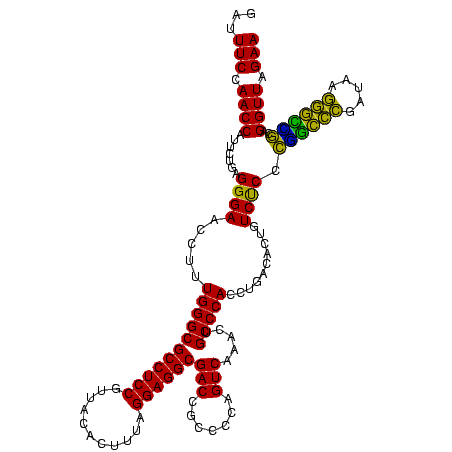
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -47.10 |
| Consensus MFE | -46.67 |
| Energy contribution | -46.67 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.26 |
| SVM RNA-class probability | 0.999981 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2280-2400 GGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAG ..(((((.....(((((...)))))((((((...........))))))))))).(((((((((..(((((((.....))))))).....((((....)))).)))))))))......... ( -47.80) >Bsubt.0 947750 120 + 4214630/2280-2400 GGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAG ..(((((.....(((((...)))))((((((...........))))))))))).(((((((((..(((((((.....))))))).....((((....)))).)))))))))......... ( -47.80) >Bcere.0 606627 120 + 5224283/2280-2400 GGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAG ..(((((.....(((((...)))))((((((...........))))))))))).(((((((((..(((((((.....))))))).....((((....)))).)))))))))......... ( -47.30) >Banth.0 534125 120 + 5227293/2280-2400 GGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAG ..(((((.....(((((...)))))((((((...........))))))))))).(((((((((..(((((((.....))))))).....((((....)))).)))))))))......... ( -47.30) >Bhalo.0 119923 120 + 4202352/2280-2400 GGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAACAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAAAAGGGAGCUUGACUGCGAG ..(((((.....(((((...)))))((((((...........))))))))))).(((((((((..(((((((.....)))))))......(((....)))..)))))))))......... ( -45.90) >Bclau.0 935920 120 + 4303871/2280-2400 GGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAACAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAGAAGGGAGCUUGACUGCGAG ..(((((.....(((((...)))))((((((...........))))))))))).((((((((((.(((((((.....)))))))......(((....)))).)))))))))......... ( -46.50) >consensus GGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAG ..(((((.....(((((...)))))((((((...........))))))))))).(((((((((..(((((((.....))))))).....((((....)))).)))))))))......... (-46.67 = -46.67 + 0.00)
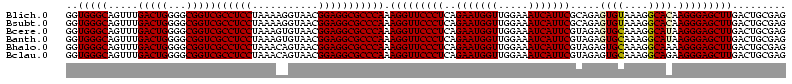
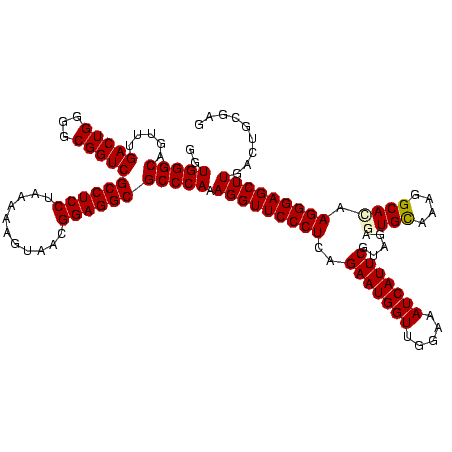
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.06 |
| Mean single sequence MFE | -37.28 |
| Consensus MFE | -35.18 |
| Energy contribution | -35.52 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.19 |
| SVM RNA-class probability | 0.999832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2280-2400 CUCGCAGUCAAGCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACC ...(((((...(((((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))).....))))))...)))))..... ( -37.40) >Bsubt.0 947750 120 + 4214630/2280-2400 CUCGCAGUCAAGCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACC ...(((((...(((((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))).....))))))...)))))..... ( -37.40) >Bcere.0 606627 120 + 5224283/2280-2400 CUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACC ...(((((...((((((((((((....)))......(((((.........)))))))))))).....(((((((((((...........))))))).....))))))...)))))..... ( -38.00) >Banth.0 534125 120 + 5227293/2280-2400 CUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACC ...(((((...((((((((((((....)))......(((((.........)))))))))))).....(((((((((((...........))))))).....))))))...)))))..... ( -38.00) >Bhalo.0 119923 120 + 4202352/2280-2400 CUCGCAGUCAAGCUCCCUUUUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACC ...(((((...((((((((.(((....)))......(((((.........))))).)))))).....(((((((((((...........))))))).....))))))...)))))..... ( -36.50) >Bclau.0 935920 120 + 4303871/2280-2400 CUCGCAGUCAAGCUCCCUUCUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACC ...(((((...((((((((.(((....)))......(((((.........))))).)))))).....(((((((((((...........))))))).....))))))...)))))..... ( -36.40) >consensus CUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACC ...(((((...((((((((.(((....)))......(((((.........))))).)))))).....(((((((((((...........))))))).....))))))...)))))..... (-35.18 = -35.52 + 0.33)

| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.11 |
| Mean single sequence MFE | -45.88 |
| Consensus MFE | -45.26 |
| Energy contribution | -45.43 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.59 |
| Structure conservation index | 0.99 |
| SVM decision value | 5.63 |
| SVM RNA-class probability | 0.999991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2320-2440 ACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAU ........((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))........ ( -47.10) >Bsubt.0 947750 120 + 4214630/2320-2440 ACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAU ........((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))........ ( -47.10) >Bcere.0 606627 120 + 5224283/2320-2440 ACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAU ........((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))........ ( -46.60) >Banth.0 534125 120 + 5227293/2320-2440 ACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGAACUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAU ........((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..((........))..)))).(((....)))))))........ ( -43.50) >Bhalo.0 119923 120 + 4202352/2320-2440 ACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAAAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAU ........((((..(((((((((..(((((((.....)))))))......(((....)))..)))))))))..((((..(((......)))..)))).(((....)))))))........ ( -45.20) >Bclau.0 935920 120 + 4303871/2320-2440 ACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAGAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAU ........((((..((((((((((.(((((((.....)))))))......(((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))........ ( -45.80) >consensus ACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAU ........((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))........ (-45.26 = -45.43 + 0.17)


| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.11 |
| Mean single sequence MFE | -32.82 |
| Consensus MFE | -30.32 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2320-2440 AUCACUAAGCCCGACUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGU ........((((((((..((..(((((.......)))))...)).))))....(((((((((......))).....(((((.........))))).))))))......))))........ ( -33.40) >Bsubt.0 947750 120 + 4214630/2320-2440 AUCACUAAGCCCGACUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGU ........((((((((..((..(((((.......)))))...)).))))....(((((((((......))).....(((((.........))))).))))))......))))........ ( -33.40) >Bcere.0 606627 120 + 5224283/2320-2440 AUCACUAAGCCCGACUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGU ........((((((((..((..(((((.......)))))...)).))))....((((((((((....)))......(((((.........))))))))))))......))))........ ( -34.00) >Banth.0 534125 120 + 5227293/2320-2440 AUCACUAAGCCCGACUUUCGUCCCUGCUCGACUUGUAGUUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGU ........((((((...........(((.((((.((.......)))))).)))((((((((((....)))......(((((.........))))))))))))....))))))........ ( -31.20) >Bhalo.0 119923 120 + 4202352/2320-2440 AUCACUAAGCCCGACUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUUUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGU ........((((((((..((..(((((.......)))))...)).))))....((((((.(((....)))......(((((.........))))).))))))......))))........ ( -32.50) >Bclau.0 935920 120 + 4303871/2320-2440 AUCACUAAGCCCGACUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUCUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGU ........((((((((..((..(((((.......)))))...)).))))....((((((.(((....)))......(((((.........))))).))))))......))))........ ( -32.40) >consensus AUCACUAAGCCCGACUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGU ........((((((((..((..(((((.......)))))...)).))))....((((((.(((....)))......(((((.........))))).))))))......))))........ (-30.32 = -30.82 + 0.50)

| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.33 |
| Mean single sequence MFE | -46.13 |
| Consensus MFE | -45.18 |
| Energy contribution | -45.23 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.98 |
| SVM decision value | 5.36 |
| SVM RNA-class probability | 0.999984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2360-2480 AUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAA (((((..(((((....(((.((...((((((..((((..(((......)))..)))).(((....)))))))))..)).(((((((....))).))))...)))..)))))..))))).. ( -46.20) >Bsubt.0 947750 120 + 4214630/2360-2480 AUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAA (((((..(((((....(((.((...((((((..((((..(((......)))..)))).(((....)))))))))..)).(((((((....))).))))...)))..)))))..))))).. ( -46.20) >Bcere.0 606627 120 + 5224283/2360-2480 AUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAA ((((((.(((((....(((......((((((..((((..(((......)))..)))).(((....))))))))).....(((((((....))).))))...)))..))))).)))))).. ( -46.10) >Banth.0 534125 120 + 5227293/2360-2480 AUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGAACUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAA ((((((.(((((....(((.((((....))))..((((.(((((((..(((((((...(((....)))..))))...)))...))))))))))).......)))..))))).)))))).. ( -45.10) >Bhalo.0 119923 120 + 4202352/2360-2480 AUUCGUAGAGUGCAAAGGCAAAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGCGGUGCCGUAUGGAAGGGCCGUCGCUCAACGGAUAA ((((((.(((((....(((......((((((..((((..(((......)))..)))).(((....))))))))).....(((((((....))).))))...)))..))))).)))))).. ( -46.60) >Bclau.0 935920 120 + 4303871/2360-2480 AUUCGUAGAGUGCAAAGGCAGAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGCGGUGCCGUAUGGAAGGGCCGUCGCUCAACGGAUAA ((((((.(((((....(((......((((((..((((..(((......)))..)))).(((....))))))))).....(((((((....))).))))...)))..))))).)))))).. ( -46.60) >consensus AUUCGUAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAA ((((((.(((((....(((......((((((..((((..(((......)))..)))).(((....))))))))).....(((((((....))).))))...)))..))))).)))))).. (-45.18 = -45.23 + 0.06)

| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.11 |
| Mean single sequence MFE | -50.13 |
| Consensus MFE | -48.60 |
| Energy contribution | -48.80 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2480-2600 AAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUG .((((..(((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).)))))(((((((....)))))))..)))...)))). ( -50.30) >Bsubt.0 947750 120 + 4214630/2480-2600 AAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUG .((((..(((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).)))))(((((((....)))))))..)))...)))). ( -50.30) >Bcere.0 606627 120 + 5224283/2480-2600 AAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUG .((((..(((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).)))))(((((((....)))))))..)))...)))). ( -50.30) >Banth.0 534125 120 + 5227293/2480-2600 AAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUG .((((..(((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).)))))(((((((....)))))))..)))...)))). ( -50.30) >Bhalo.0 119923 120 + 4202352/2480-2600 AAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUG .(((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).)))))((((((......)))))).)))))..)))). ( -49.40) >Bclau.0 935920 120 + 4303871/2480-2600 AAGCUACCCUGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUG .(((((((((.(((((..((((((.((((((((...((.......))...))))))))..))).((((((((.(((....)))))))..)))))))......))))).)))))..)))). ( -50.20) >consensus AAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUG .((((((((..(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).)))))(((((((....)))))))..))))..)))). (-48.60 = -48.80 + 0.20)

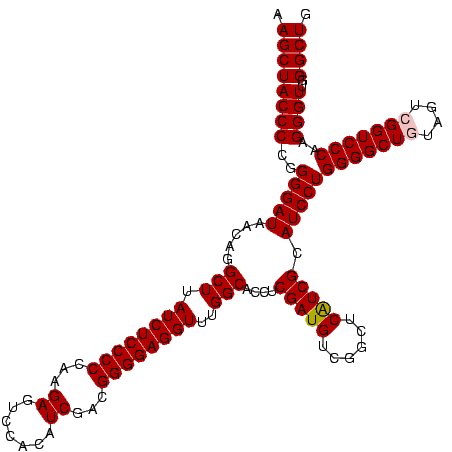
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.11 |
| Mean single sequence MFE | -43.83 |
| Consensus MFE | -43.02 |
| Energy contribution | -42.80 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.732975 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2480-2600 CAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGACUCUUGGGGGAGAUAAGCCUGUUAUCCCCGGGGUAGCUU .(((...(((((.((((..(((.....((((((((.((.....))((((...(((((.....)))))...))))........)))))))).((......))))).)))).))))).))). ( -43.70) >Bsubt.0 947750 120 + 4214630/2480-2600 CAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGACUCUUGGGGGAGAUAAGCCUGUUAUCCCCGGGGUAGCUU .(((...(((((.((((..(((.....((((((((.((.....))((((...(((((.....)))))...))))........)))))))).((......))))).)))).))))).))). ( -43.70) >Bcere.0 606627 120 + 5224283/2480-2600 CAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGACUCUUGGGGGAGAUAAGCCUGUUAUCCCCGGGGUAGCUU .(((...(((((.((((..(((.....((((((((.((.....))((((...(((((.....)))))...))))........)))))))).((......))))).)))).))))).))). ( -43.70) >Banth.0 534125 120 + 5227293/2480-2600 CAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGACUCUUGGGGGAGAUAAGCCUGUUAUCCCCGGGGUAGCUU .(((...(((((.((((..(((.....((((((((.((.....))((((...(((((.....)))))...))))........)))))))).((......))))).)))).))))).))). ( -43.70) >Bhalo.0 119923 120 + 4202352/2480-2600 CAGCCCAACCCUUGGGACCUACUUCAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGAACUCUUGGGGGAGAUAAGCCUGUUAUCCCCAGGGUAGCUU .(((...(((((.(((...........((((((((.((.....))((((...(((((.....)))))...))))........))))))))..(((((.....))))))))))))).))). ( -43.10) >Bclau.0 935920 120 + 4303871/2480-2600 CAGCCCAACCCUUGGGACCUACUUCAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGACUCUUGGGGGAGAUAAGCCUGUUAUCCCCAGGGUAGCUU .(((...(((((.(((...........((((((((..(.(((...((((...(((((.....)))))...)))).))).)..))))))))..(((((.....))))))))))))).))). ( -45.10) >consensus CAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGGACUCUUGGGGGAGAUAAGCCUGUUAUCCCCGGGGUAGCUU .(((...(((((.((((..........((((((((.((.....))((((...(((((.....)))))...))))........))))))))..((((....)))).)))).))))).))). (-43.02 = -42.80 + -0.22)

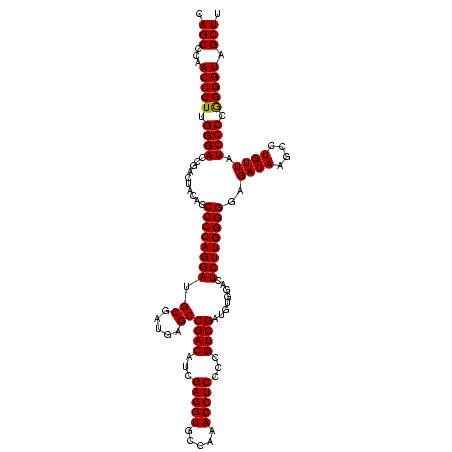
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.28 |
| Mean single sequence MFE | -51.08 |
| Consensus MFE | -50.62 |
| Energy contribution | -50.53 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2520-2640 CCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGU (((...(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........... ( -51.70) >Bsubt.0 947750 120 + 4214630/2520-2640 CCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGU (((...(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........... ( -51.70) >Bcere.0 606627 120 + 5224283/2520-2640 CCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGU (((...(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........... ( -51.70) >Banth.0 534125 120 + 5227293/2520-2640 CCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGU (((...(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........... ( -51.70) >Bhalo.0 119923 120 + 4202352/2520-2640 UCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGU .....((((((..(((((.....)))))..))))))...((((((((((((((((......)))))).)))))(((((.....))))).....)))))..(((..(((....)))..))) ( -47.90) >Bclau.0 935920 120 + 4303871/2520-2640 CCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCGUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGU (((...(((((..(((((.....)))))..)))))((((((((((((((((((((......)))))).)))))(((((.....))))).....)))....)))))))))........... ( -51.80) >consensus CCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGU (((...(((((..(((((.....)))))..)))))(((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))........... (-50.62 = -50.53 + -0.08)

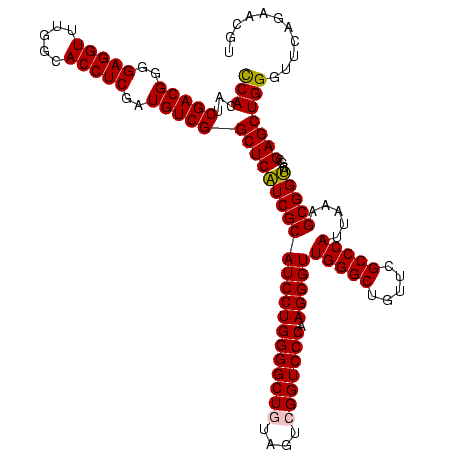
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.28 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -41.03 |
| Energy contribution | -40.92 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -0.92 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888459 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2520-2640 ACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGG ...........((((((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))...))) ( -41.50) >Bsubt.0 947750 120 + 4214630/2520-2640 ACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGG ...........((((((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))...))) ( -41.50) >Bcere.0 606627 120 + 5224283/2520-2640 ACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGG ...........((((((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))...))) ( -41.50) >Banth.0 534125 120 + 5227293/2520-2640 ACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGG ...........((((((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))...))) ( -41.50) >Bhalo.0 119923 120 + 4202352/2520-2640 ACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCUACUUCAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGA ((((..........(((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...)))))))).. ( -40.50) >Bclau.0 935920 120 + 4303871/2520-2640 ACGUUCUGAACCCAGCUCGCGUGCCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCUACUUCAGCCCCAGGAUGCGACGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGG ...........(((((((((((..........(((((.....)))))...((((((.(........).))))))..))).)))))((((...(((((.....)))))...))))...))) ( -42.50) >consensus ACGUUCUGAACCCAGCUCGCGUACCGCUUUAAUGGGCGAACAGCCCAACCCUUGGGACCGACUACAGCCCCAGGAUGCGAUGAGCCGACAUCGAGGUGCCAAACCUCCCCGUCGAUGUGG ...........((((((((((((.........(((((.....)))))...((((((.(........).)))))).)))).)))))((((...(((((.....)))))...))))...))) (-41.03 = -40.92 + -0.11)

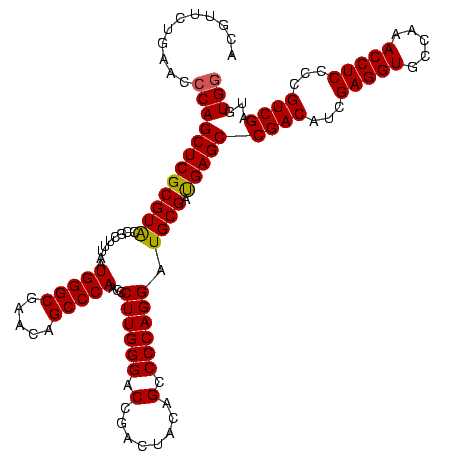
| Location | 922,209 – 922,329 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.94 |
| Mean single sequence MFE | -52.92 |
| Consensus MFE | -49.19 |
| Energy contribution | -52.17 |
| Covariance contribution | 2.97 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 120 + 4222334/2560-2680 UCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGCAGGAA .(((((((((((((((....))))))).)))))(((((.....))))).....)))(..((((((.((((..(.(((((((....))).)))).)..))))..).)))))..)....... ( -54.00) >Bsubt.0 947750 120 + 4214630/2560-2680 UCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGCAGGAA .(((((((((((((((....))))))).)))))(((((.....))))).....)))(..((((((.((((..(.(((((((....))).)))).)..))))..).)))))..)....... ( -54.00) >Bcere.0 606627 120 + 5224283/2560-2680 UCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGGAA .(((((((((((((((....))))))).)))))(((((.....))))).....)))(..((((((.((((..(.(((((((....))).)))).)..))))..).)))))..)....... ( -52.10) >Banth.0 534125 120 + 5227293/2560-2680 UCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGUGGGCGUAGGAA .(((((((((((((((....))))))).)))))(((((.....))))).....)))(..((((((.((((..(.(((((((....))).)))).)..))))..).)))))..)....... ( -52.10) >Bhalo.0 119923 120 + 4202352/2560-2680 UCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGUAGGAA .((((((((((((((......)))))).)))))(((((.....))))).....)))(..((((((.((((..(.(((((((....))).)))).)..))))..).)))))..)....... ( -52.10) >Bclau.0 935920 120 + 4303871/2560-2680 UCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGCACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGUAGGAA .((((((((((((((......)))))).)))))(((((.....))))).....)))((.((((((.((((..(.(((((((....))).)))).)..))))..).))))).))....... ( -53.20) >consensus UCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGCGGGCGUAGGAA .(((((((((((((((....))))))).)))))(((((.....))))).....)))((.((((((.((((..(.(((((((....))).)))).)..))))..).))))).))....... (-49.19 = -52.17 + 2.97)
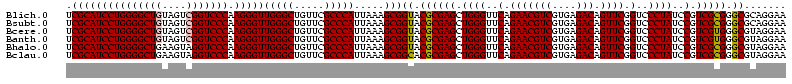
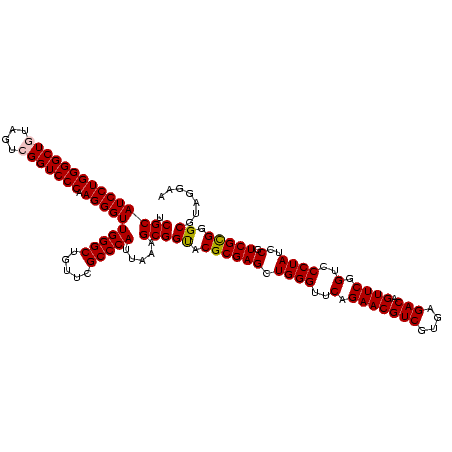
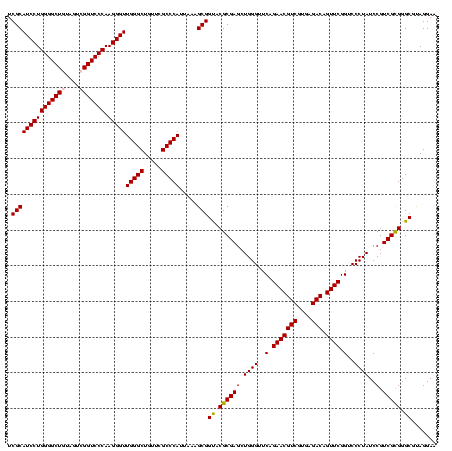
| Location | 922,209 – 922,328 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.91 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -30.67 |
| Energy contribution | -27.60 |
| Covariance contribution | -3.07 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 922209 119 + 4222334/2826-2946 GAGAUUUCCCAUUCCUAUAAG-GAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACC ......((((.((((.....)-)))....(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))).)))....... ( -33.10) >Bsubt.0 947750 118 + 4214630/2826-2946 GAGAUUUCCCAUUCC-GCAAG-GAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGGCUUAACC .......(((.((((-....)-)))....(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))..)))....... ( -38.20) >Bcere.0 606627 118 + 5224283/2826-2946 GAGAUUUCCCAUAGC-GUAAG-CUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC ......((((.((((-....)-)))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).)))....... ( -35.50) >Banth.0 534125 118 + 5227293/2826-2946 GAGAUUUCCCAUAGC-GUAAG-CUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC ......((((.((((-....)-)))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).)))....... ( -35.50) >Bhalo.0 119923 119 + 4202352/2826-2946 GAGAUUUCCCAUGGA-GUAAAUCCAGUAAGACCCCUUAGAGAUGAUGAGGUUGAUAGGUCUGGUGUGGAAGCGUGGCGACACGUGAAGCUGACAGAUACUAAUCGGUCGAGGACUUAUCC ..(((.((((.((((-.....))))....(((((((((.......)))))..((((((((((..((....(((((....)))))...))...))))).)).)))))))).)))...))). ( -35.80) >Bclau.0 935920 118 + 4303871/2826-2946 GAGAUUUCCCAUGGC-GUAAG-CCAGUAAGACCCCUUAGAGAUGAUGAGGUUGAUAGGUCAGAAGUGGAAGCGUGGCGACACGUGGAGCGGACUGAUACUAAUCGGUCGAGGACUUAUCC ...........((((-....)-)))........(((((.......)))))..((((((((....((....(((((....)))))...)).(((((((....)))))))...)))))))). ( -40.00) >consensus GAGAUUUCCCAUAGC_GUAAG_CAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGCGUGGCGACACGUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACC ......((((.(((........)))....(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...))))).)))....... (-30.67 = -27.60 + -3.07)
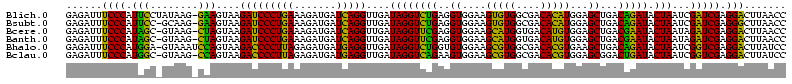
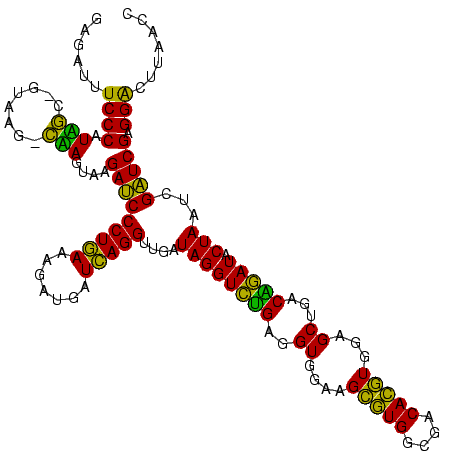
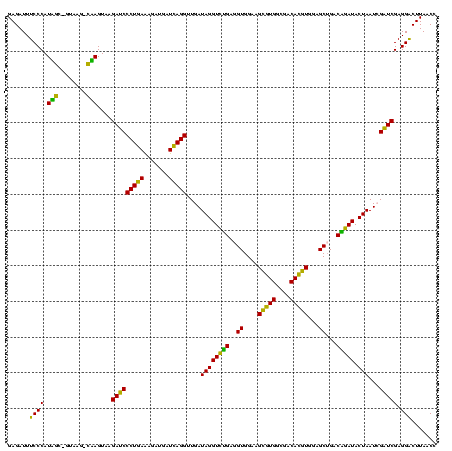
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:18:30 2006