
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 3,363,369 – 3,363,480 |
| Length | 111 |
| Max. P | 0.999441 |

| Location | 3,363,369 – 3,363,477 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.14 |
| Mean single sequence MFE | -29.76 |
| Consensus MFE | -11.30 |
| Energy contribution | -10.98 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.55 |
| Mean z-score | -3.03 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988132 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
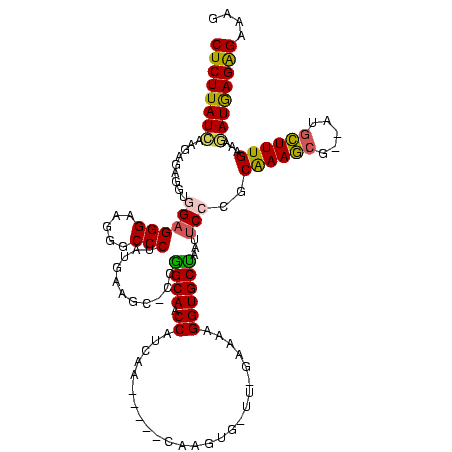
>Bsubt.0 3363369 108 + 4214630/0-120 AAGAAAAGCCUUUCUGCACACUUGAGCAGAAAGGCUUAUAUAAAAU----GCCUUUCUCUCAUCUUUCAAAGCUGAACGCUUCGUGUGAAUUGGCACCAUUUC-AA-CA-GUG-----UU ..((((.(((......(((((.((((.((((((((...........----)))))))))))).......((((.....)))).)))))....)))....))))-..-..-...-----.. ( -30.70) >Blich.0 3302282 113 + 4222334/0-120 GAAAAAAGCCUUUCUGCACACAUGAGCAGAAAGGCUUUGGAAAAGUCUGGGCCUUUCUCUCAUCUUUCAAAGCAUAACGCUUCGCGUGAAUUGGCACCAUUUC-AA-CACGUG-----UU (((((((((((((((((........)))))))))))))(((((.((....)).)))))......)))).((((.....))))((((((((.((....)).)))-..-.)))))-----.. ( -35.40) >Banth.0 4073923 106 + 5227293/0-120 AUAAAAACCCCCUCUUCAC-UUUGAAGAGGGGAACAU---AAUUGU-----UCCUCCUCUUAUUAUGCAAAACAUU-CGUUUUGCAGGUAUUAGCACCGUUUC-AAACAUUUCGU---UU ..........(((((((..-...)))))))((((((.---...)))-----)))...........((((((((...-.))))))))(((......))).....-...........---.. ( -29.90) >Bhalo.0 3590655 113 + 4202352/0-120 CAAAAAAACCUUUCCGC---CAUGAGCAGAAAGGAAUGUUUUACGUCU--UCCCUUCUCUCAUCUCUCAAAGCAU--CGCUUUGCAGGAAUUAGCACCUUUUCCAAGCGAUUUGCUGCUU ...............((---.(((((.((((.((((((.....)))..--))).))))))))).......(((((--(((((....((((..........))))))))))..)))))).. ( -25.20) >Bclau.0 2740129 106 - 4303871/0-120 UGAAAAACACCCGUUCCGGGAUUGGAAAGGGUGUUGUUUUCACGAAAU---GAAUCCCCUUAUCUGCCAAAGUU---CCCUUUGCUGGAUUUAGCACCUUUUC--AUCGCAA------UG ((((((((((((.(((((....))))).))))))..)))))).(..((---(((....((.((((..(((((..---..)))))..))))..))......)))--))..)..------.. ( -27.60) >consensus AAAAAAACCCUUUCUGCAC_CUUGAGCAGAAAGGCAUUUUAAAAGU_U___CCCUUCUCUCAUCUUUCAAAGCAU__CGCUUUGCAGGAAUUAGCACCAUUUC_AA_CACGUG_____UU ......................((((.((((((((...............))))))))))))((...((((((.....))))))...))............................... (-11.30 = -10.98 + -0.32)

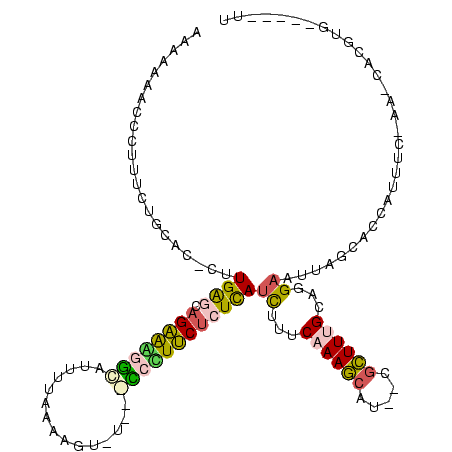
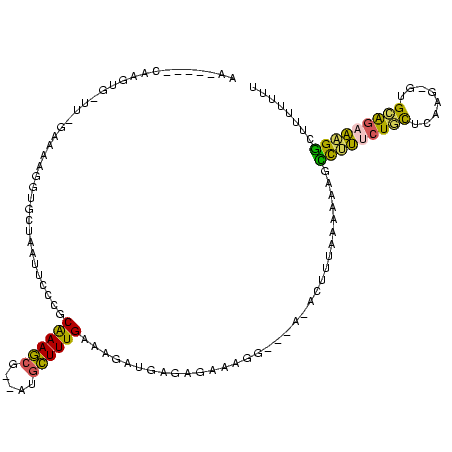
| Location | 3,363,369 – 3,363,477 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 58.14 |
| Mean single sequence MFE | -33.00 |
| Consensus MFE | -14.80 |
| Energy contribution | -13.48 |
| Covariance contribution | -1.32 |
| Combinations/Pair | 1.67 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.995569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 3363369 108 + 4214630/0-120 AA-----CAC-UG-UU-GAAAUGGUGCCAAUUCACACGAAGCGUUCAGCUUUGAAAGAUGAGAGAAAGGC----AUUUUAUAUAAGCCUUUCUGCUCAAGUGUGCAGAAAGGCUUUUCUU ..-----...-..-..-(((..((((((..(((.((((((((.....)))))).....)).)))...)))----)))......((((((((((((.(....).))))))))))))))).. ( -33.70) >Blich.0 3302282 113 + 4222334/0-120 AA-----CACGUG-UU-GAAAUGGUGCCAAUUCACGCGAAGCGUUAUGCUUUGAAAGAUGAGAGAAAGGCCCAGACUUUUCCAAAGCCUUUCUGCUCAUGUGUGCAGAAAGGCUUUUUUC ..-----....((-..-(((.(((.(((..(((((.((((((.....))))))...).)))).....))))))...)))..))((((((((((((.(....).))))))))))))..... ( -36.60) >Banth.0 4073923 106 + 5227293/0-120 AA---ACGAAAUGUUU-GAAACGGUGCUAAUACCUGCAAAACG-AAUGUUUUGCAUAAUAAGAGGAGGA-----ACAAUU---AUGUUCCCCUCUUCAAA-GUGAAGAGGGGGUUUUUAU ((---((....(((((-.....((((....))))((((((((.-...))))))))............))-----)))...---.....(((((((((...-..))))))))))))).... ( -31.90) >Bhalo.0 3590655 113 + 4202352/0-120 AAGCAGCAAAUCGCUUGGAAAAGGUGCUAAUUCCUGCAAAGCG--AUGCUUUGAGAGAUGAGAGAAGGGA--AGACGUAAAACAUUCCUUUCUGCUCAUG---GCGGAAAGGUUUUUUUG ((((.....(((((((.(...(((........))).).)))))--)).((((..(..(((((((((((((--(...........))))))))).))))).---.)..))))))))..... ( -32.90) >Bclau.0 2740129 106 - 4303871/0-120 CA------UUGCGAU--GAAAAGGUGCUAAAUCCAGCAAAGGG---AACUUUGGCAGAUAAGGGGAUUC---AUUUCGUGAAAACAACACCCUUUCCAAUCCCGGAACGGGUGUUUUUCA ..------.....((--((((.(((.((..(((...(((((..---..)))))...)))...)).))).---.))))))(((((..((((((.((((......)))).))))))))))). ( -29.90) >consensus AA_____CAAGUG_UU_GAAAAGGUGCUAAUUCCCGCAAAGCG__AUGCUUUGAAAGAUGAGAGAAAGG___A_ACUUUAAAAAAGCCUUUCUGCUCAAG_GUGCAGAAAGGCUUUUUUU ....................................((((((.....)))))).................................(((((((((........)))))))))........ (-14.80 = -13.48 + -1.32)

| Location | 3,363,369 – 3,363,480 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.39 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -23.81 |
| Energy contribution | -23.13 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.64 |
| SVM decision value | 3.61 |
| SVM RNA-class probability | 0.999441 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 3363369 111 + 4214630/52-172 CUCUUAUCAAGAGAGGUGGAGGGAAGUGCCCUAUGAAGC-CCGGCAACCAUCAA-----CAC-UG-UU-GAAAUGGUGCCAAUUCACACGAAGCGUUCAGCUUUGAAAGAUGAGAGAAAG ((((((((..(((.(((..((((.....)))).....))-).((((.(((((((-----(..-.)-))-))..)))))))..)))...((((((.....))))))...)))))))).... ( -40.00) >Blich.0 3302282 112 + 4222334/52-172 CUCUUAUCCAGAGAGGUGGAGGGAAGUGCCCUAUGAAAC-CCGGCAACCAUCAA-----CACGUG-UU-GAAAUGGUGCCAAUUCACGCGAAGCGUUAUGCUUUGAAAGAUGAGAGAAAG ((((((((..(((.(((..((((.....)))).....))-).((((.(((((((-----(....)-))-))..)))))))..)))...((((((.....))))))...)))))))).... ( -39.30) >Banth.0 4073923 114 + 5227293/52-172 CUCUUAUUGAGAGCGGUGGAGGGAAAGGCCCUGUGAAAC-CCGGCAACCUUCAA---ACGAAAUGUUU-GAAACGGUGCUAAUACCUGCAAAACG-AAUGUUUUGCAUAAUAAGAGGAGG (((((((((.....((((.((((.....)))).......-..((((.(((((((---((.....))))-)))..))))))..))))((((((((.-...))))))))))))))))).... ( -37.70) >Bhalo.0 3590655 117 + 4202352/52-172 CUCUUAUCAUGAGAGGUGGAGGGACUGGCCCGAUGAAGC-CCAGCAACCGCCAAGCAGCAAAUCGCUUGGAAAAGGUGCUAAUUCCUGCAAAGCG--AUGCUUUGAGAGAUGAGAGAAGG ((((((((.........((((((.((..........)))-))((((.((.((((((........))))))....))))))...)))..((((((.--..))))))...)))))))).... ( -40.10) >Bclau.0 2740129 109 - 4303871/52-172 CUCUUAUAACGAGAAGCGGAGGGACUGGCCCAAUGAAGCUUCAGCAACCAUUCA------UUGCGAU--GAAAAGGUGCUAAAUCCAGCAAAGGG---AACUUUGGCAGAUAAGGGGAUU (((((((.....(((((...(((.....)))......)))))((((.((.((((------(....))--)))..))))))........(((((..---..)))))....))))))).... ( -29.90) >consensus CUCUUAUCAAGAGAGGUGGAGGGAAGGGCCCUAUGAAGC_CCGGCAACCAUCAA_____CAAGUG_UU_GAAAAGGUGCUAAUUCCCGCAAAGCG__AUGCUUUGAAAGAUGAGAGAAAG ((((((((.........((((((.....)))...........((((.((.........................))))))...)))..((((((.....))))))...)))))))).... (-23.81 = -23.13 + -0.68)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:46 2006