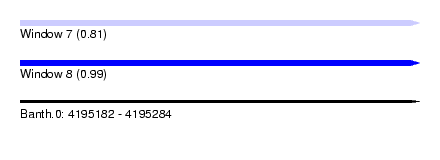
| Sequence ID | Banth.0 |
|---|---|
| Location | 4,195,182 – 4,195,284 |
| Length | 102 |
| Max. P | 0.987387 |

| Location | 4,195,182 – 4,195,284 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.47 |
| Mean single sequence MFE | -26.59 |
| Consensus MFE | -6.89 |
| Energy contribution | -6.67 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.26 |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.810908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
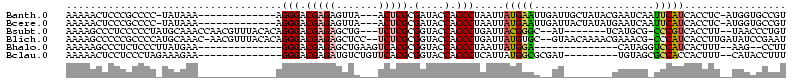
>Banth.0 4195182 102 + 5227293/0-120 AAAAACUCCCGCCCC-UAUAAA-------------AGGGACGAGAGUUA---ACUCGCGAUACCACCCUAAUUAUGAAUUGAUUGCUAUACGAAUCAAUUCAUCACCUC-AUGGUGCCGU ...(((((.((.(((-(.....-------------)))).)).))))).---....(((.(((((........(((((((((((........)))))))))))......-.))))).))) ( -30.86) >Bcere.0 4142216 102 + 5224283/0-120 AAAAACUCCCGCCCC-UAUAAA-------------AGGGACGAGAGUUA---ACUCGCGAUACCACCCUAAUUAUGAAUUGAUUACUAUAUGAAUCAAUUCAUCACCUC-AUGGUGCCGU ...(((((.((.(((-(.....-------------)))).)).))))).---....(((.(((((........(((((((((((........)))))))))))......-.))))).))) ( -30.76) >Bsubt.0 2800084 105 + 4214630/0-120 AAAAGCCCUCCCCCUAUGCAAACCAACGUUUACACAGGGACGAGAGCUG---UCUCGCGGUACCACCCUGAUUACGGGC--AU-------UCAUGCG-CCCGUCACCUU--UAACCCUGU ...(((.(((.((((.((.((((....)))).)).))))..))).))).---..(((.((.....)).)))..((((((--..-------......)-)))))......--......... ( -28.10) >Blich.0 2757871 114 + 4222334/0-120 AAAAGCCCCCGCCCCAUGCAAAC-AACGUUUACACAGGGACGAGAGCUCC--UCUCGCGGUACCACCCUGAUUAUGUGC--GUAACAAAACGAAACG-CCCAUCACCUUGAUAUCCGAAU ..........((.....))....-..((..(((((((((.((((((...)--))))).(....).)))))...(((.((--((...........)))-).))).....)).))..))... ( -21.90) >Bhalo.0 2839289 88 - 4202352/0-120 AAAAAGCCCUCUCCCUUAUGAA--------------GGGACGAGAGCUGAAGUCACGCGGUACCACCCUAAUUAUGGA--------------CAUAGGUCCAUCACUUU--AAG--CCUU ....(((.((((((((.....)--------------)))).))).)))(((((.....((.....))......(((((--------------(....)))))).)))))--...--.... ( -23.90) >Bclau.0 2586831 95 - 4303871/0-120 AAAAACUCCUCCCUAGAAAGAA--------------GGGACGAGAUGUCUGUUCACGCGGUACCACCCUCAUUAUGGCGCGAU---------UGUAGCGCCACCACUUU--CAUACCUUU .....(((.(((((.......)--------------)))).)))..((((((....)))).))...........((((((...---------....)))))).......--......... ( -24.00) >consensus AAAAACCCCCCCCCC_UAUAAA_____________AGGGACGAGAGCUA___UCUCGCGGUACCACCCUAAUUAUGGAC__AU________GAAUCG_UCCAUCACCUU__UAGCCCCGU ....................................(((.(((((.......))))).(....).))).....(((((....................)))))................. ( -6.89 = -6.67 + -0.22)
| Location | 4,195,182 – 4,195,284 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.47 |
| Mean single sequence MFE | -39.40 |
| Consensus MFE | -13.81 |
| Energy contribution | -12.98 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.55 |
| Mean z-score | -3.66 |
| Structure conservation index | 0.35 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
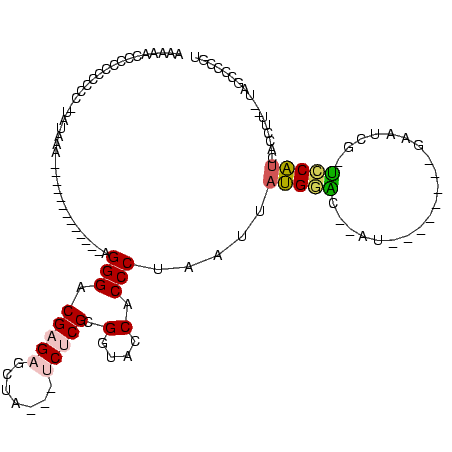
>Banth.0 4195182 102 + 5227293/0-120 ACGGCACCAU-GAGGUGAUGAAUUGAUUCGUAUAGCAAUCAAUUCAUAAUUAGGGUGGUAUCGCGAGU---UAACUCUCGUCCCU-------------UUUAUA-GGGGCGGGAGUUUUU .((..(((((-..(((.((((((((((((.....).))))))))))).)))...)))))..)).....---.(((((((((((((-------------.....)-))))))))))))... ( -40.90) >Bcere.0 4142216 102 + 5224283/0-120 ACGGCACCAU-GAGGUGAUGAAUUGAUUCAUAUAGUAAUCAAUUCAUAAUUAGGGUGGUAUCGCGAGU---UAACUCUCGUCCCU-------------UUUAUA-GGGGCGGGAGUUUUU .((..(((((-..(((.((((((((((((.....).))))))))))).)))...)))))..)).....---.(((((((((((((-------------.....)-))))))))))))... ( -40.20) >Bsubt.0 2800084 105 + 4214630/0-120 ACAGGGUUA--AAGGUGACGGG-CGCAUGA-------AU--GCCCGUAAUCAGGGUGGUACCGCGAGA---CAGCUCUCGUCCCUGUGUAAACGUUGGUUUGCAUAGGGGGAGGGCUUUU .....(((.--..(((.(((((-((.....-------.)--)))))).)))...(((....)))..))---)(((((((.(((((((((((((....))))))))))))))))))))... ( -47.80) >Blich.0 2757871 114 + 4222334/0-120 AUUCGGAUAUCAAGGUGAUGGG-CGUUUCGUUUUGUUAC--GCACAUAAUCAGGGUGGUACCGCGAGA--GGAGCUCUCGUCCCUGUGUAAACGUU-GUUUGCAUGGGGCGGGGGCUUUU ...(((.(((((...((((.((-(((..(.....)..))--)).)...))))...))))))))....(--((((((((((((((.((((((((...-))))))))))))))))))))))) ( -46.60) >Bhalo.0 2839289 88 - 4202352/0-120 AAGG--CUU--AAAGUGAUGGACCUAUG--------------UCCAUAAUUAGGGUGGUACCGCGUGACUUCAGCUCUCGUCCC--------------UUCAUAAGGGAGAGGGCUUUUU ....--(((--((....((((((....)--------------)))))..)))))(((....)))........(((((((.((((--------------(.....)))))))))))).... ( -29.90) >Bclau.0 2586831 95 - 4303871/0-120 AAAGGUAUG--AAAGUGGUGGCGCUACA---------AUCGCGCCAUAAUGAGGGUGGUACCGCGUGAACAGACAUCUCGUCCC--------------UUCUUUCUAGGGAGGAGUUUUU ...(((((.--(..((.(((((((....---------...))))))).)).....).)))))..............(((.((((--------------(.......))))).)))..... ( -31.00) >consensus ACGGGACUA__AAGGUGAUGGA_CGAUUC________AU__AUCCAUAAUUAGGGUGGUACCGCGAGA___CAGCUCUCGUCCCU_____________UUUAUA_GGGGCGGGAGUUUUU .............(((.(((((....................))))).)))...(((....)))........(((((((.((((.....................)))).)))))))... (-13.81 = -12.98 + -0.83)

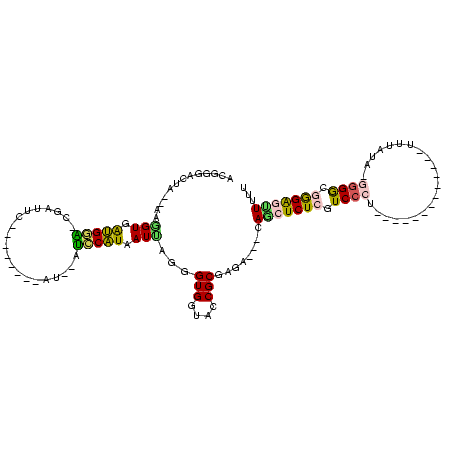
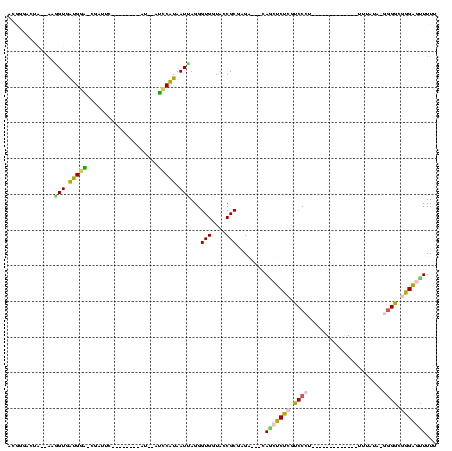
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:14:53 2006