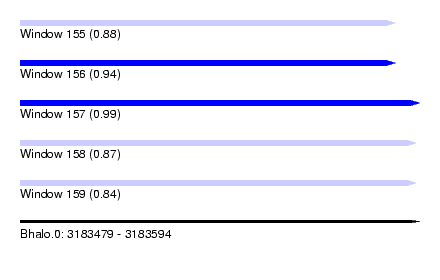
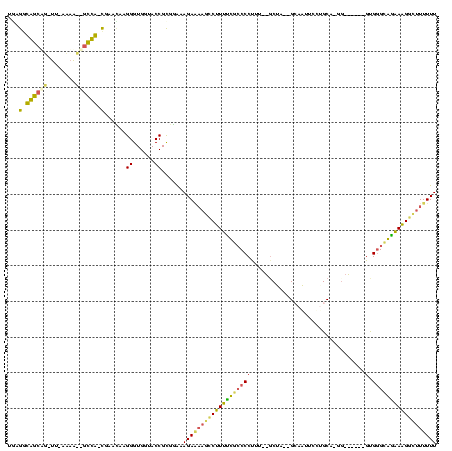
| Sequence ID | Bhalo.0 |
|---|---|
| Location | 3,183,479 – 3,183,594 |
| Length | 115 |
| Max. P | 0.993339 |

| Location | 3,183,479 – 3,183,587 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.77 |
| Mean single sequence MFE | -38.22 |
| Consensus MFE | -12.70 |
| Energy contribution | -15.57 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.97 |
| Structure conservation index | 0.33 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.883208 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3183479 108 + 4202352/0-120 AAAAAAAUCUUUUUACCCAC------GC-UGCCAGAAUUUC--UAGC--AAAGGGGCAAAAAGAUUUCUCUUUUCACGGUACCACCCUCAUUCG-UAGCAUUUUUACAAAUGCUACCUCA ....((((((((((.(((.(------..-(((.((.....)--).))--)..)))).))))))))))..........((.....)).......(-((((((((....))))))))).... ( -28.30) >Bclau.0 2774870 102 + 4303871/0-120 AAAAAAGACUUUCUGCGCGC------CCAUGCUGGGAU--C--CAGC--AAGGGGCGAGAAAG-----UCUUCACGCGGUACCACCCUUUUUCGCUAACAAUGGU-UAGCCUCUAGCAAG ....((((((((((.(((.(------((.((((((...--)--))))--).))))))))))))-----))))...((((.....)).......((((((....))-)))).....))... ( -42.80) >Bsubt.0 2896109 117 + 4214630/0-120 AAAAAAGCCCUUCCGCCCACAAUCAGCCGCGCAGUAACUGCGAUAGCUAAAAGGGGCGAAAAGGCUUUUCUUUCCGCGGUACCACCCUUGUUUA-CGGC--CUGUUAAUCGGCCGUCUCA ..(((((((.((.(((((......(((..(((((...)))))...))).....))))).)).)))))))........((.....)).......(-((((--(........)))))).... ( -39.30) >Blich.0 2850042 112 + 4222334/0-120 AAAAAAGCCUUCCCGCCCACUGUUGACGCUGCACACCUUGC---AGGCCAAAGGGGCGAAAAGGCUUUUCUUUCCGCGGUACCACCCUUGUUCG-CGGC--UUUUGAA--AGCCGCCUUA ..(((((((((..(((((.((.(((.(.(((((.....)))---))).))))))))))..)))))))))........((.....)).......(-((((--((....)--)))))).... ( -42.50) >consensus AAAAAAGCCUUUCCGCCCAC______CC_UGCAGGAAUUGC__UAGC__AAAGGGGCAAAAAGGCUUUUCUUUCCGCGGUACCACCCUUGUUCG_CAGC__UUGU_AA_CAGCCACCUCA ..((((((((((((((((............(((.....)))............))))))))))))))))........((.....)).........((((............))))..... (-12.70 = -15.57 + 2.88)

| Location | 3,183,479 – 3,183,587 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 59.77 |
| Mean single sequence MFE | -50.80 |
| Consensus MFE | -23.05 |
| Energy contribution | -22.55 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.54 |
| Mean z-score | -5.62 |
| Structure conservation index | 0.45 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938827 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3183479 108 + 4202352/0-120 UGAGGUAGCAUUUGUAAAAAUGCUA-CGAAUGAGGGUGGUACCGUGAAAAGAGAAAUCUUUUUGCCCCUUU--GCUA--GAAAUUCUGGCA-GC------GUGGGUAAAAAGAUUUUUUU ....(((((((((....))))))))-)......((......))......(((((((((((((((((((.((--((((--(.....))))))-).------).)))))))))))))))))) ( -44.60) >Bclau.0 2774870 102 + 4303871/0-120 CUUGCUAGAGGCUA-ACCAUUGUUAGCGAAAAAGGGUGGUACCGCGUGAAGA-----CUUUCUCGCCCCUU--GCUG--G--AUCCCAGCAUGG------GCGCGCAGAAAGUCUUUUUU ...((.....((((-((....))))))......((......))))..(((((-----((((((((((((.(--((((--(--...)))))).))------).))).)))))))))))... ( -44.80) >Bsubt.0 2896109 117 + 4214630/0-120 UGAGACGGCCGAUUAACAG--GCCG-UAAACAAGGGUGGUACCGCGGAAAGAAAAGCCUUUUCGCCCCUUUUAGCUAUCGCAGUUACUGCGCGGCUGAUUGUGGGCGGAAGGGCUUUUUU ....((((((........)--))))-)........(((....)))....(((((((((((((((((((..((((((..(((((...))))).))))))..).)))))))))))))))))) ( -57.30) >Blich.0 2850042 112 + 4222334/0-120 UAAGGCGGCU--UUCAAAA--GCCG-CGAACAAGGGUGGUACCGCGGAAAGAAAAGCCUUUUCGCCCCUUUGGCCU---GCAAGGUGUGCAGCGUCAACAGUGGGCGGGAAGGCUUUUUU ....((((((--(....))--))))-)........(((....)))....(((((((((((..((((((((((((((---(((.....))))).))))).)).)))))..))))))))))) ( -56.50) >consensus UGAGGCAGCAG_UU_AAAA__GCCA_CGAACAAGGGUGGUACCGCGGAAAGAAAAGCCUUUUCGCCCCUUU__GCUA__GCAAUUCCUGCA_GG______GUGGGCAGAAAGGCUUUUUU ...(.((((((........)))))).)......((......))......(((((((((((((((((((................................).)))))))))))))))))) (-23.05 = -22.55 + -0.50)

| Location | 3,183,479 – 3,183,594 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.06 |
| Mean single sequence MFE | -37.88 |
| Consensus MFE | -17.01 |
| Energy contribution | -16.82 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993339 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3183479 115 + 4202352/40-160 CACUACACCCGUUAUCAAAAUGAGACGAUUUGAUCGUCCAUGAGGUAGCAUUUGUAAAAAUGCUA-CGAAUGAGGGUGGUACCGUGAAAAGAGAAAUCUUUUUGCCCCUUU--GCUA--G .....(((((....((...(((.((((((...)))))))))...(((((((((....))))))))-)))....))))).....(..((((((....))))))..)......--....--. ( -39.30) >Bclau.0 2774870 108 + 4303871/40-160 GAACGCAUUCGUUAUAAAAAC--UGCAAGCCGGAUUUCGGCUUGCUAGAGGCUA-ACCAUUGUUAGCGAAAAAGGGUGGUACCGCGUGAAGA-----CUUUCUCGCCCCUU--GCUG--G ....((.(((..........(--(((((((((.....)))))))).))..((((-((....))))))))).(((((.(....)(((.(((..-----..))).))))))))--))..--. ( -35.40) >Bsubt.0 2896109 112 + 4214630/40-160 -GAUUCACUCGUUACUAAAGC--GGAUAG--AAAUAUCCAUGAGACGGCCGAUUAACAG--GCCG-UAAACAAGGGUGGUACCGCGGAAAGAAAAGCCUUUUCGCCCCUUUUAGCUAUCG -......((((((.....)))--(((((.--...)))))..)))((((((........)--))))-)....(((((.(....)((((((((......))))))))))))).......... ( -37.10) >Blich.0 2850042 110 + 4222334/40-160 AGGUUCACUCGUUAUUAAACC--GGAUAAGCGAUUAUCCAUAAGGCGGCU--UUCAAAA--GCCG-CGAACAAGGGUGGUACCGCGGAAAGAAAAGCCUUUUCGCCCCUUUGGCCU---G .(((((((((...........--((((((....)))))).....((((((--(....))--))))-)......)))))).)))((((((((......))))))))...........---. ( -39.70) >consensus _AAUUCACUCGUUAUUAAAAC__GGAUAG_CGAAUAUCCAUGAGGCAGCAG_UU_AAAA__GCCA_CGAACAAGGGUGGUACCGCGGAAAGAAAAGCCUUUUCGCCCCUUU__GCUA__G .....(((((.............(((((......)))))....(.((((((........)))))).)......))))).....((((((((......))))))))............... (-17.01 = -16.82 + -0.19)

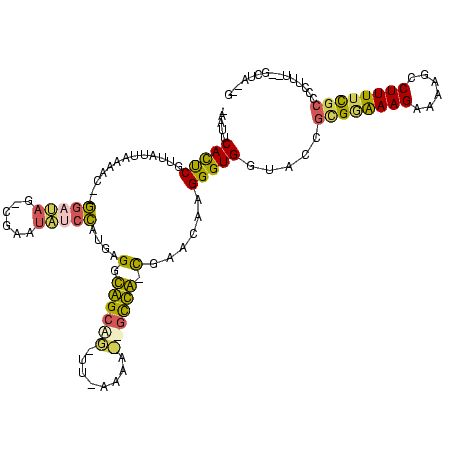
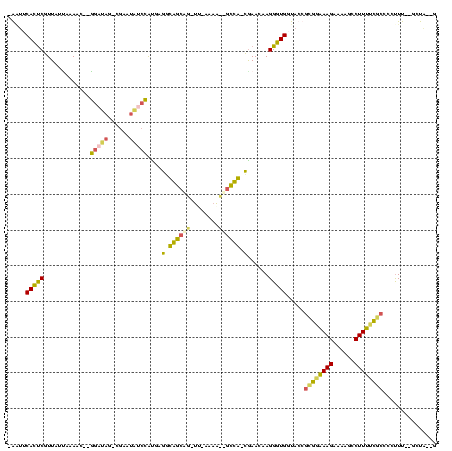
| Location | 3,183,479 – 3,183,593 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 53.87 |
| Mean single sequence MFE | -36.25 |
| Consensus MFE | -12.15 |
| Energy contribution | -11.71 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.34 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.874319 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3183479 114 + 4202352/80-200 CAGCCUGCUUUAGUAGAUG-UUGCCGGGUGGUUUUU----CACUACACCCGUUAUCAAAAUGAGACGAUUUGAUCGUCCAUGAGGUAGCAUUUGUAAAAAUGCUA-CGAAUGAGGGUGGU ((((((((....))))..)-)))...(((((....)----)))).(((((....((...(((.((((((...)))))))))...(((((((((....))))))))-)))....))))).. ( -38.10) >Bclau.0 2774870 102 + 4303871/80-200 -------UUGUAGUAAGAG--CGCCGGAUG--CUGU----GAACGCAUUCGUUAUAAAAAC--UGCAAGCCGGAUUUCGGCUUGCUAGAGGCUA-ACCAUUGUUAGCGAAAAAGGGUGGU -------............--(((((((((--(.(.----...)))))))..........(--(((((((((.....)))))))).))..((((-((....)))))).......)))).. ( -35.20) >Bsubt.0 2896109 101 + 4214630/80-200 -----AUG-ACAGUAGGACUUGGCCGGGUGAACUU------GAUUCACUCGUUACUAAAGC--GGAUAG--AAAUAUCCAUGAGACGGCCGAUUAACAG--GCCG-UAAACAAGGGUGGU -----...-.........((((..((((((((...------..))))))))..........--(((((.--...))))).....((((((........)--))))-)...))))...... ( -32.10) >Blich.0 2850042 110 + 4222334/80-200 --UCCAUGUUUAGUAGGGC-UGGCCGGGUGAACAUUACGAAGGUUCACUCGUUAUUAAACC--GGAUAAGCGAUUAUCCAUAAGGCGGCU--UUCAAAA--GCCG-CGAACAAGGGUGGU --.((((.((((((((((.-...))((((((((.........)))))))).))))))))((--((((((....)))))).....((((((--(....))--))))-)......)))))). ( -39.60) >consensus _____AUGUUUAGUAGGAC_UGGCCGGGUGAACUUU_____AAUUCACUCGUUAUUAAAAC__GGAUAG_CGAAUAUCCAUGAGGCAGCAG_UU_AAAA__GCCA_CGAACAAGGGUGGU ......................((((((((...............))))).............(((((......)))))....(.((((((........)))))).).......)))... (-12.15 = -11.71 + -0.44)

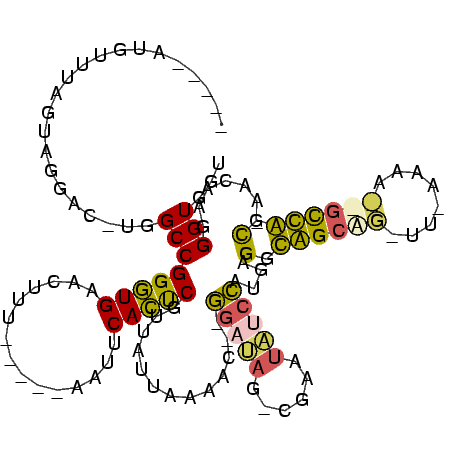
| Location | 3,183,479 – 3,183,593 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 56.18 |
| Mean single sequence MFE | -30.07 |
| Consensus MFE | -17.05 |
| Energy contribution | -15.30 |
| Covariance contribution | -1.75 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.838833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3183479 114 + 4202352/120-240 UG-AAGCUCACCUCGGAGUGCGACUUCGAACAAUCGGGCACAGCCUGCUUUAGUAGAUG-UUGCCGGGUGGUUUUU----CACUACACCCGUUAUCAAAAUGAGACGAUUUGAUCGUCCA ((-((((.....((((((.....)))))).......((((((..((((....)))).))-.))))(((((......----.....)))))))).)))...((.((((((...)))))))) ( -33.60) >Bclau.0 2774870 94 + 4303871/120-240 UG-AAACUCACCUCUGAGUGCGUCUCUGAACA---------------UUGUAGUAAGAG--CGCCGGAUG--CUGU----GAACGCAUUCGUUAUAAAAAC--UGCAAGCCGGAUUUCGG ..-..(((((....))))).((..((((....---------------(((((((.....--...((((((--(.(.----...))))))))........))--)))))..))))...)). ( -24.09) >Bsubt.0 2896109 94 + 4214630/120-240 GGUGAACUCGCCUCAGAGUGCCAGUCUGAA---------------AUG-ACAGUAGGACUUGGCCGGGUGAACUU------GAUUCACUCGUUACUAAAGC--GGAUAG--AAAUAUCCA (((((((((......))))((((((((...---------------...-......)))).))))((((((((...------..))))))))))))).....--(((((.--...))))). ( -31.60) >Blich.0 2850042 110 + 4222334/120-240 UGUGAACUCCCCUCUGAGUGUCAGUCUGAACGAUU-------UCCAUGUUUAGUAGGGC-UGGCCGGGUGAACAUUACGAAGGUUCACUCGUUAUUAAACC--GGAUAAGCGAUUAUCCA .....((((......))))(((((((((((((...-------....)))))....))))-))))(((((((((.........)))))))))..........--((((((....)))))). ( -31.00) >consensus UG_AAACUCACCUCUGAGUGCCACUCUGAACA_____________AUGUUUAGUAGGAC_UGGCCGGGUGAACUUU_____AAUUCACUCGUUAUUAAAAC__GGAUAG_CGAAUAUCCA .....((((......))))((((((((............................)))).))))((((((...............))))))............(((((......))))). (-17.05 = -15.30 + -1.75)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:37 2006