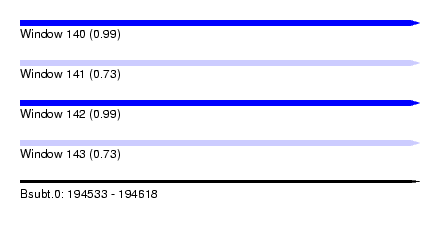
| Sequence ID | Bsubt.0 |
|---|---|
| Location | 194,533 – 194,618 |
| Length | 85 |
| Max. P | 0.993369 |

| Location | 194,533 – 194,618 |
|---|---|
| Length | 85 |
| Sequences | 3 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -30.12 |
| Energy contribution | -29.90 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993369 |
| Prediction | RNA |
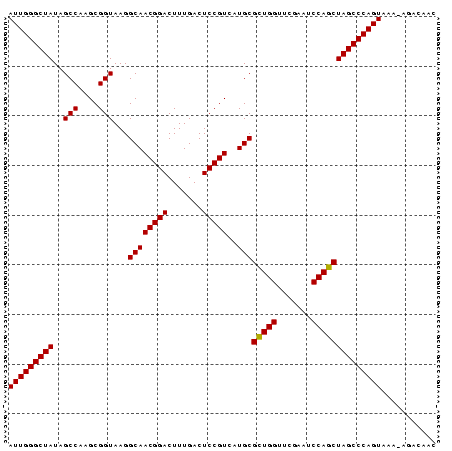
Download alignment: ClustalW | MAF
>Bsubt.0 194533 85 + 4214630 AUUGGGCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUCAUGCGUUGGUUCGAAUCCAGCUAGCCCAGUCAC-AGACACC (((((((((..(((....)))...((((((((.......)))))..)))(((((.......))))))))))))))...-....... ( -28.10) >Bhalo.0 2952399 85 + 4202352 AUUGGGCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUGAUGCGCUGGUUCGAAUCCAGCUAGCCCAGUAAA-AAAAAAC (((((((((..(((....)))...((((((((.......)))))..)))(((((.......))))))))))))))...-....... ( -31.10) >Blich.0 176970 86 + 4222334 AUUGGGCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUCAUGCGCUGGUUCGAAUCCAGCUAGCCCAGUUUUUGGACAUC (((((((((..(((....)))...((((((((.......)))))..)))(((((.......))))))))))))))........... ( -30.50) >consensus AUUGGGCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUCAUGCGCUGGUUCGAAUCCAGCUAGCCCAGUAAA_AGACAAC (((((((((..(((....)))...((((((((.......)))))..)))(((((.......))))))))))))))........... (-30.12 = -29.90 + -0.22)

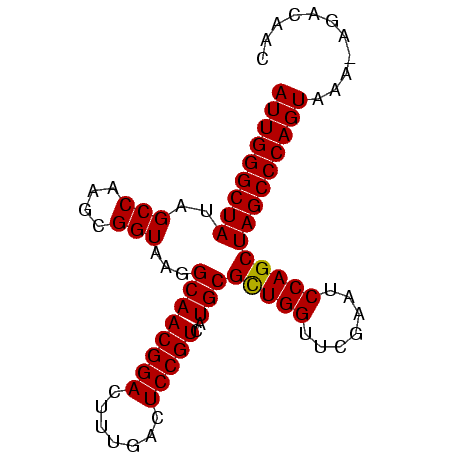
| Location | 194,533 – 194,618 |
|---|---|
| Length | 85 |
| Sequences | 3 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -26.07 |
| Energy contribution | -26.73 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 194533 85 + 4214630 GGUGUCU-GUGACUGGGCUAGCUGGAUUCGAACCAACGCAUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCAAU ...(((.-..)))((((((((((((.......)))..))..((((((.......))))))(((.........)))..))))))).. ( -24.00) >Bhalo.0 2952399 85 + 4202352 GUUUUUU-UUUACUGGGCUAGCUGGAUUCGAACCAGCGCAUCACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCAAU .......-.....((((((((((((.......))))).....(((((.......))))).(((.........)))..))))))).. ( -27.50) >Blich.0 176970 86 + 4222334 GAUGUCCAAAAACUGGGCUAGCUGGAUUCGAACCAGCGCAUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCAAU .............((((((((((((.......)))))....((((((.......))))))(((.........)))..))))))).. ( -29.30) >consensus GAUGUCU_AUAACUGGGCUAGCUGGAUUCGAACCAGCGCAUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCAAU .............((((((((((((.......)))))....((((((.......))))))(((.........)))..))))))).. (-26.07 = -26.73 + 0.67)

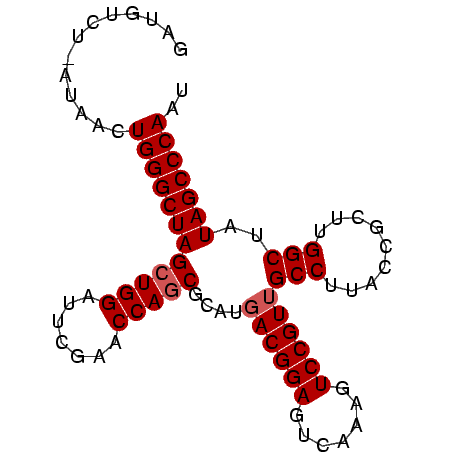

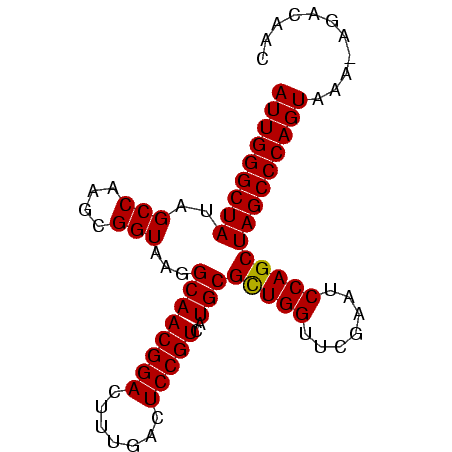
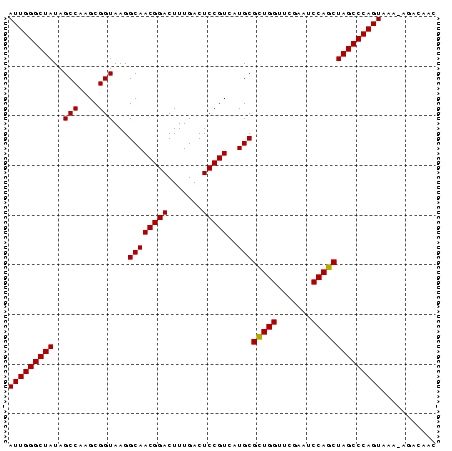
| Location | 194,533 – 194,618 |
|---|---|
| Length | 85 |
| Sequences | 3 |
| Columns | 86 |
| Reading direction | forward |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -30.12 |
| Energy contribution | -29.90 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.46 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.993369 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 194533 85 + 4214630/0-86 AUUGGGCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUCAUGCGUUGGUUCGAAUCCAGCUAGCCCAGUCAC-AGACACC (((((((((..(((....)))...((((((((.......)))))..)))(((((.......))))))))))))))...-....... ( -28.10) >Bhalo.0 2952399 85 + 4202352/0-86 AUUGGGCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUGAUGCGCUGGUUCGAAUCCAGCUAGCCCAGUAAA-AAAAAAC (((((((((..(((....)))...((((((((.......)))))..)))(((((.......))))))))))))))...-....... ( -31.10) >Blich.0 176970 86 + 4222334/0-86 AUUGGGCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUCAUGCGCUGGUUCGAAUCCAGCUAGCCCAGUUUUUGGACAUC (((((((((..(((....)))...((((((((.......)))))..)))(((((.......))))))))))))))........... ( -30.50) >consensus AUUGGGCUAUAGCCAAGCGGUAAGGCAACGGACUUUGACUCCGUCAUGCGCUGGUUCGAAUCCAGCUAGCCCAGUAAA_AGACAAC (((((((((..(((....)))...((((((((.......)))))..)))(((((.......))))))))))))))........... (-30.12 = -29.90 + -0.22)

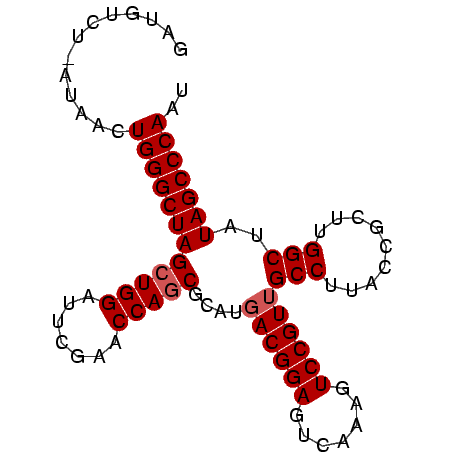
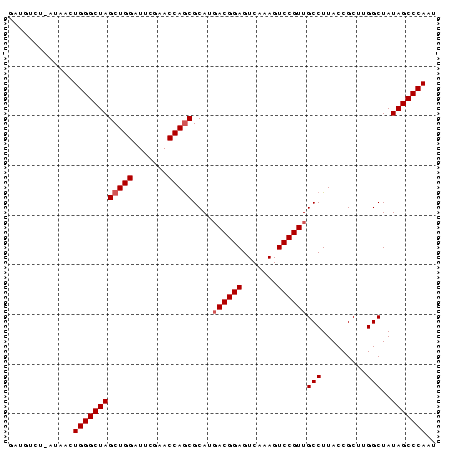
| Location | 194,533 – 194,618 |
|---|---|
| Length | 85 |
| Sequences | 3 |
| Columns | 86 |
| Reading direction | reverse |
| Mean pairwise identity | 91.05 |
| Mean single sequence MFE | -26.93 |
| Consensus MFE | -26.07 |
| Energy contribution | -26.73 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734558 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bsubt.0 194533 85 + 4214630/0-86 GGUGUCU-GUGACUGGGCUAGCUGGAUUCGAACCAACGCAUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCAAU ...(((.-..)))((((((((((((.......)))..))..((((((.......))))))(((.........)))..))))))).. ( -24.00) >Bhalo.0 2952399 85 + 4202352/0-86 GUUUUUU-UUUACUGGGCUAGCUGGAUUCGAACCAGCGCAUCACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCAAU .......-.....((((((((((((.......))))).....(((((.......))))).(((.........)))..))))))).. ( -27.50) >Blich.0 176970 86 + 4222334/0-86 GAUGUCCAAAAACUGGGCUAGCUGGAUUCGAACCAGCGCAUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCAAU .............((((((((((((.......)))))....((((((.......))))))(((.........)))..))))))).. ( -29.30) >consensus GAUGUCU_AUAACUGGGCUAGCUGGAUUCGAACCAGCGCAUGACGGAGUCAAAGUCCGUUGCCUUACCGCUUGGCUAUAGCCCAAU .............((((((((((((.......)))))....((((((.......))))))(((.........)))..))))))).. (-26.07 = -26.73 + 0.67)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:21 2006