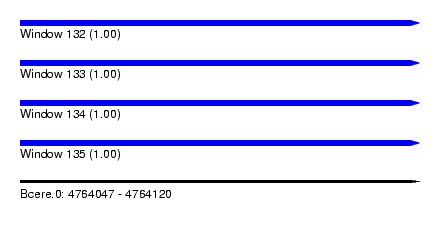
| Sequence ID | Bcere.0 |
|---|---|
| Location | 4,764,047 – 4,764,120 |
| Length | 73 |
| Max. P | 0.999905 |

| Location | 4,764,047 – 4,764,120 |
|---|---|
| Length | 73 |
| Sequences | 3 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.96 |
| Structure conservation index | 0.80 |
| SVM decision value | 4.45 |
| SVM RNA-class probability | 0.999900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
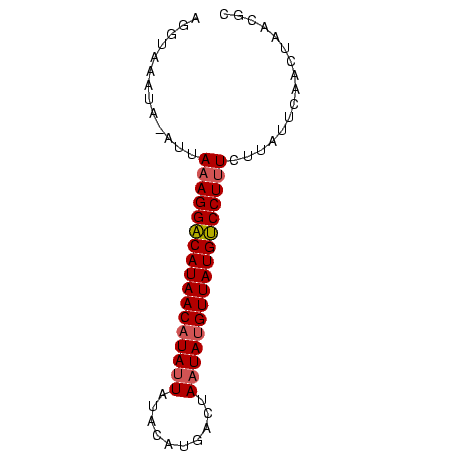
>Bcere.0 4764047 73 + 5224283 AGGUCAAUACAUGAAAGGGCAUAACAUAUUAGUCAUAUAUAAUAUGUUAUGCCCUUUGUUAUUCAACUCACGC .............((((((((((((((((((........))))))))))))))))))................ ( -25.40) >Bhalo.0 307207 73 + 4202352 AGGUAAAUAAAUUAAAGGACAUAACAUAGUUUACAUGGCUAAUAGGUUAUGUCCUUUUUUGUUCAACUAACGC .(((.((((((..((((((((((((.(((((.....)))))....))))))))))))))))))..)))..... ( -20.60) >Bclau.0 2264084 72 - 4303871 AGUUAGAUA-AUUAAAGGACAUAACAUAUUGUAUAUGACGAAUAUGUUAUGUCCUCUCUCACUCAGCGAACAC .(((.((..-.....(((((((((((((((((.....)).))))))))))))))).......)))))...... ( -21.04) >consensus AGGUAAAUA_AUUAAAGGACAUAACAUAUUAUACAUGACUAAUAUGUUAUGUCCUUUCUUAUUCAACUAACGC .............(((((((((((((((((..........)))))))))))))))))................ (-17.94 = -18.50 + 0.56)

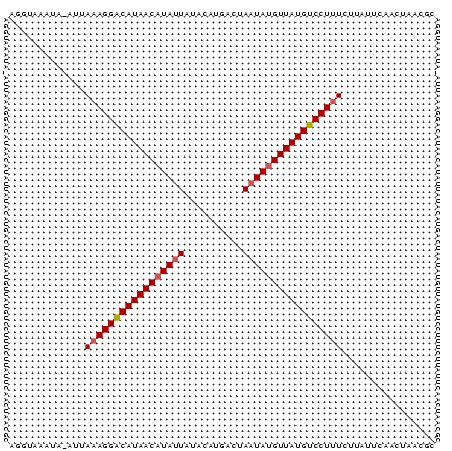
| Location | 4,764,047 – 4,764,120 |
|---|---|
| Length | 73 |
| Sequences | 3 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
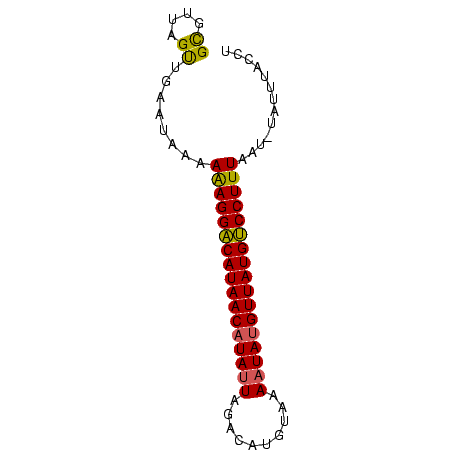
>Bcere.0 4764047 73 + 5224283 GCGUGAGUUGAAUAACAAAGGGCAUAACAUAUUAUAUAUGACUAAUAUGUUAUGCCCUUUCAUGUAUUGACCU (((((((((....))).(((((((((((((((((........)))))))))))))))))))))))........ ( -28.50) >Bhalo.0 307207 73 + 4202352 GCGUUAGUUGAACAAAAAAGGACAUAACCUAUUAGCCAUGUAAACUAUGUUAUGUCCUUUAAUUUAUUUACCU ..(((.....)))...((((((((((((....(((.........))).))))))))))))............. ( -14.80) >Bclau.0 2264084 72 - 4303871 GUGUUCGCUGAGUGAGAGAGGACAUAACAUAUUCGUCAUAUACAAUAUGUUAUGUCCUUUAAU-UAUCUAACU ...(((((...)))))(((((((((((((((((.((.....)))))))))))))))))))...-......... ( -23.20) >consensus GCGUUAGUUGAAUAAAAAAGGACAUAACAUAUUAGACAUGUAAAAUAUGUUAUGUCCUUUAAU_UAUUUACCU ((....))........(((((((((((((((((..........)))))))))))))))))............. (-19.44 = -19.00 + -0.44)

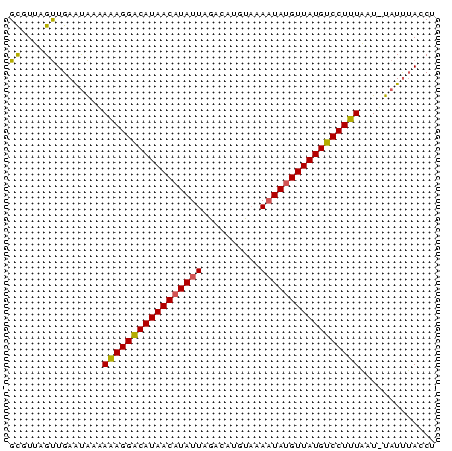
| Location | 4,764,047 – 4,764,120 |
|---|---|
| Length | 73 |
| Sequences | 3 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -22.35 |
| Consensus MFE | -17.94 |
| Energy contribution | -18.50 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.06 |
| Mean z-score | -4.96 |
| Structure conservation index | 0.80 |
| SVM decision value | 4.45 |
| SVM RNA-class probability | 0.999900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 4764047 73 + 5224283/0-73 AGGUCAAUACAUGAAAGGGCAUAACAUAUUAGUCAUAUAUAAUAUGUUAUGCCCUUUGUUAUUCAACUCACGC .............((((((((((((((((((........))))))))))))))))))................ ( -25.40) >Bhalo.0 307207 73 + 4202352/0-73 AGGUAAAUAAAUUAAAGGACAUAACAUAGUUUACAUGGCUAAUAGGUUAUGUCCUUUUUUGUUCAACUAACGC .(((.((((((..((((((((((((.(((((.....)))))....))))))))))))))))))..)))..... ( -20.60) >Bclau.0 2264084 72 - 4303871/0-73 AGUUAGAUA-AUUAAAGGACAUAACAUAUUGUAUAUGACGAAUAUGUUAUGUCCUCUCUCACUCAGCGAACAC .(((.((..-.....(((((((((((((((((.....)).))))))))))))))).......)))))...... ( -21.04) >consensus AGGUAAAUA_AUUAAAGGACAUAACAUAUUAUACAUGACUAAUAUGUUAUGUCCUUUCUUAUUCAACUAACGC .............(((((((((((((((((..........)))))))))))))))))................ (-17.94 = -18.50 + 0.56)

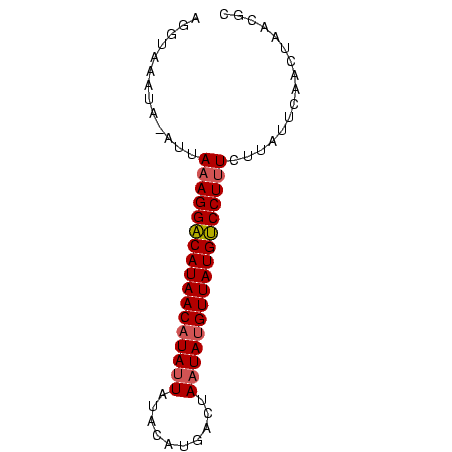
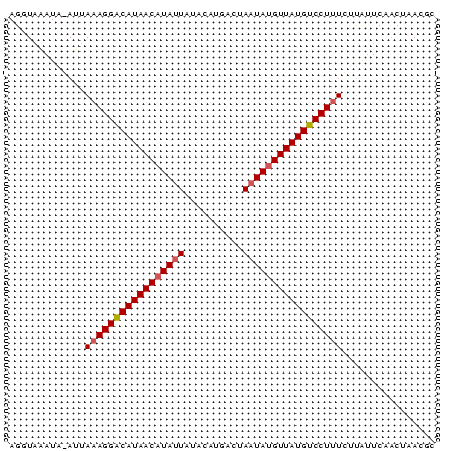
| Location | 4,764,047 – 4,764,120 |
|---|---|
| Length | 73 |
| Sequences | 3 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 74.43 |
| Mean single sequence MFE | -22.17 |
| Consensus MFE | -19.44 |
| Energy contribution | -19.00 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.21 |
| Mean z-score | -4.55 |
| Structure conservation index | 0.88 |
| SVM decision value | 4.48 |
| SVM RNA-class probability | 0.999905 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 4764047 73 + 5224283/0-73 GCGUGAGUUGAAUAACAAAGGGCAUAACAUAUUAUAUAUGACUAAUAUGUUAUGCCCUUUCAUGUAUUGACCU (((((((((....))).(((((((((((((((((........)))))))))))))))))))))))........ ( -28.50) >Bhalo.0 307207 73 + 4202352/0-73 GCGUUAGUUGAACAAAAAAGGACAUAACCUAUUAGCCAUGUAAACUAUGUUAUGUCCUUUAAUUUAUUUACCU ..(((.....)))...((((((((((((....(((.........))).))))))))))))............. ( -14.80) >Bclau.0 2264084 72 - 4303871/0-73 GUGUUCGCUGAGUGAGAGAGGACAUAACAUAUUCGUCAUAUACAAUAUGUUAUGUCCUUUAAU-UAUCUAACU ...(((((...)))))(((((((((((((((((.((.....)))))))))))))))))))...-......... ( -23.20) >consensus GCGUUAGUUGAAUAAAAAAGGACAUAACAUAUUAGACAUGUAAAAUAUGUUAUGUCCUUUAAU_UAUUUACCU ((....))........(((((((((((((((((..........)))))))))))))))))............. (-19.44 = -19.00 + -0.44)

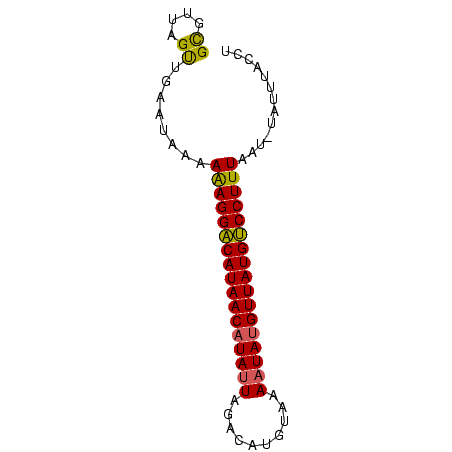
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:12 2006