
| Sequence ID | Bhalo.0 |
|---|---|
| Location | 3,177,403 – 3,177,521 |
| Length | 118 |
| Max. P | 0.997708 |

| Location | 3,177,403 – 3,177,509 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.04 |
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -20.89 |
| Energy contribution | -19.64 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.70 |
| SVM decision value | 2.91 |
| SVM RNA-class probability | 0.997708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
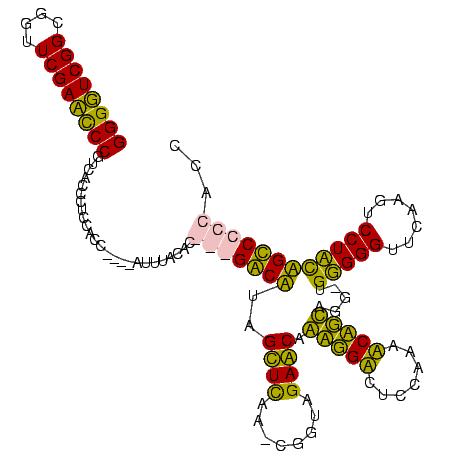
>Bhalo.0 3177403 106 - 4202352/40-160 AGUCGCCACACUGA----------AAUGAU--AAAUUGGCAGGGGCAGUAGGAAUCGAACCCACA-CCGGAGGUUUUGGAGACCUCUGUUCUACCG-UUAAACUAUGCCCCUAUGAAAAU ....((((......----------......--....))))(((((((((.((........)))).-.((((((((.....))))))))........-........)))))))........ ( -30.29) >Bclau.0 2856206 118 - 4303871/40-160 AACCGCCAUAUUGAUACAUGCUUAAACGAUUCAAGUUGGCAGGGGCAGUAGGAAUCGAACCCACA-CUGGAGGUUUUGGAGACCUCUGUUCUACCG-UUAAACUAUGCCCCUAUGAAAAU ....((((..((((................))))..))))(((((((.(((.(((.((((.....-..(((((((.....)))))))))))....)-))...))))))))))........ ( -32.70) >Bcere.0 4650670 96 + 5224283/40-160 AACCGCCAUAAUUU---------------------CUGGUGCCGACUAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUACCAAUUGAGCUAAGUC---GGCUAAAU ....(((.....((---------------------(((((....)))))))((((((............((((((.....)))))).((((........))))))))))---)))..... ( -28.20) >Banth.0 4651692 96 + 5227293/40-160 AACCGCCAUGAUUU---------------------CUGGUGCCGACUAGAGGACUUGAACCCCCAACCUACUGAUUACAAGUCAGUUGCUCUCCCAAUUGAGCUAAGUC---GGCUAAAU ....(((.....((---------------------(((((....)))))))((((((............((((((.....)))))).((((........))))))))))---)))..... ( -28.20) >consensus AACCGCCAUAAUGA_____________________CUGGCACCGACAAGAGGAAUCGAACCCACA_CCGAAGGAUUACAAGACAGCUGCUCUACCA_UUAAACUAAGCC___AGCAAAAU ....((((............................))))(((((((...((........))......(((((((.....)))))))((((........))))..)))))))........ (-20.89 = -19.64 + -1.25)

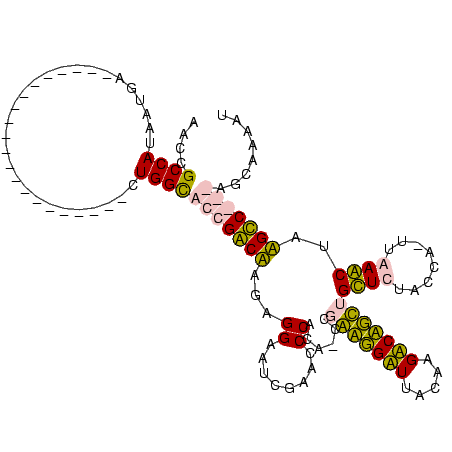
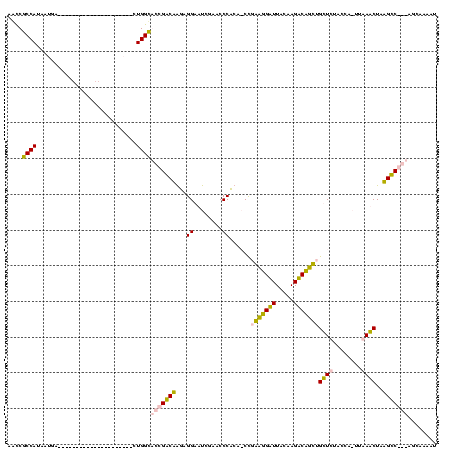
| Location | 3,177,403 – 3,177,509 |
|---|---|
| Length | 106 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.04 |
| Mean single sequence MFE | -29.83 |
| Consensus MFE | -24.64 |
| Energy contribution | -21.39 |
| Covariance contribution | -3.25 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3177403 106 - 4202352/40-160 AUUUUCAUAGGGGCAUAGUUUAA-CGGUAGAACAGAGGUCUCCAAAACCUCCGG-UGUGGGUUCGAUUCCUACUGCCCCUGCCAAUUU--AUCAUU----------UCAGUGUGGCGACU ........(((((((..((((..-.....)))).(((((.......)))))...-.(((((.......))))))))))))((((....--..(((.----------...))))))).... ( -28.80) >Bclau.0 2856206 118 - 4303871/40-160 AUUUUCAUAGGGGCAUAGUUUAA-CGGUAGAACAGAGGUCUCCAAAACCUCCAG-UGUGGGUUCGAUUCCUACUGCCCCUGCCAACUUGAAUCGUUUAAGCAUGUAUCAAUAUGGCGGUU ........(((((((..((((..-.....)))).(((((.......)))))...-.(((((.......))))))))))))((((..((((..(((......)))..))))..)))).... ( -32.90) >Bcere.0 4650670 96 + 5224283/40-160 AUUUAGCC---GACUUAGCUCAAUUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUAGUCGGCACCAG---------------------AAAUUAUGGCGGUU .....(((---(((...((((........)))).(((((.......)))))....((((((.......)))))))))))).(((.---------------------......)))..... ( -28.80) >Banth.0 4651692 96 + 5227293/40-160 AUUUAGCC---GACUUAGCUCAAUUGGGAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUAGUCGGCACCAG---------------------AAAUCAUGGCGGUU .....(((---((((.((.......((((((((..(..(((((.......)))))..)..))))...))))))))))))).(((.---------------------......)))..... ( -28.81) >consensus AUUUACAC___GACAUAGCUCAA_CGGUAGAACAAAGGACUCCAAAACAGCAGG_UGGGGGUUCAAGUCCUACAGCCCCCACCAA_____________________AAAUUAUGGCGGUU .........((((((..((((........)))).(((((.......))))).....(((((.......)))))))))))(((((............................)))))... (-24.64 = -21.39 + -3.25)

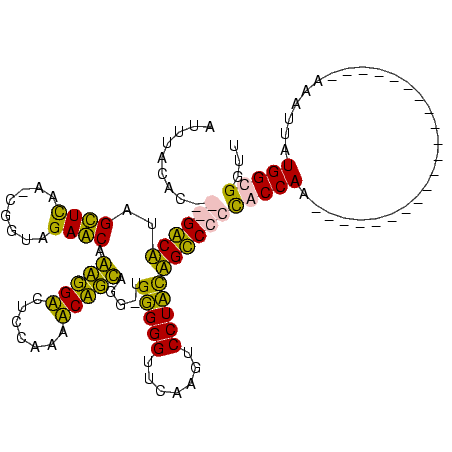
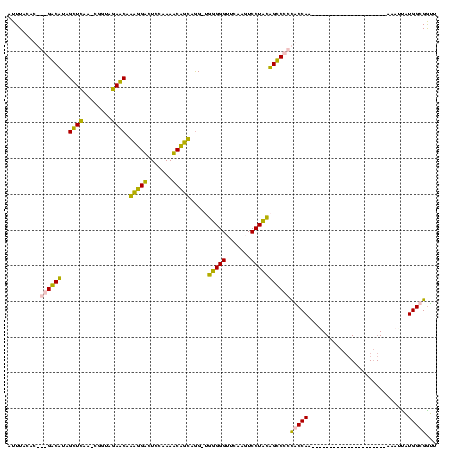
| Location | 3,177,403 – 3,177,521 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.97 |
| Mean single sequence MFE | -37.95 |
| Consensus MFE | -34.75 |
| Energy contribution | -31.00 |
| Covariance contribution | -3.75 |
| Combinations/Pair | 1.41 |
| Mean z-score | -0.98 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.970392 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bhalo.0 3177403 118 - 4202352/77-197 GGGUUCGGCGGUUCGAAUCCGUCCCCCUCCACCAUUUAUUUUCAUAGGGGCAUAGUUUAA-CGGUAGAACAGAGGUCUCCAAAACCUCCGG-UGUGGGUUCGAUUCCUACUGCCCCUGCC (((...(((((.......))))).))).................((((((((..((((..-.....)))).(((((.......)))))...-.(((((.......))))))))))))).. ( -36.00) >Bclau.0 2856206 114 - 4303871/77-197 GGGUUCGGCGGUUCGAAUCCGUCCCCCUCCACC----AUUUUCAUAGGGGCAUAGUUUAA-CGGUAGAACAGAGGUCUCCAAAACCUCCAG-UGUGGGUUCGAUUCCUACUGCCCCUGCC (((...(((((.......))))).)))......----.......((((((((..((((..-.....)))).(((((.......)))))...-.(((((.......))))))))))))).. ( -36.00) >Bcere.0 4650670 113 + 5224283/77-197 GGGGUCGGCGGUUCGAUCCCGUCAUCCUCCACC----AUUUAGCC---GACUUAGCUCAAUUGGUAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUAGUCGGCACC ((((..(((((.......)))))..)).))...----.....(((---(((...((((........)))).(((((.......)))))....((((((.......))))))))))))... ( -39.90) >Banth.0 4651692 113 + 5227293/77-197 GGGGUCGGCGGUUCGAUCCCGUCAUCCUCCACC----AUUUAGCC---GACUUAGCUCAAUUGGGAGAGCAACUGACUUGUAAUCAGUAGGUUGGGGGUUCAAGUCCUCUAGUCGGCACC ((((..(((((.......)))))..)).))...----.....(((---((((.((.......((((((((..(..(((((.......)))))..)..))))...)))))))))))))... ( -39.91) >consensus GGGGUCGGCGGUUCGAACCCGUCACCCUCCACC____AUUUACAC___GACAUAGCUCAA_CGGUAGAACAAAGGACUCCAAAACAGCAGG_UGGGGGUUCAAGUCCUACAGCCCCCACC ((((((((....)))))))).........................(((((((..((((........)))).(((((.......))))).....(((((.......))))))))))))... (-34.75 = -31.00 + -3.75)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:17:08 2006