
| Sequence ID | Banth.0 |
|---|---|
| Location | 4,131,099 – 4,131,150 |
| Length | 51 |
| Max. P | 0.974993 |

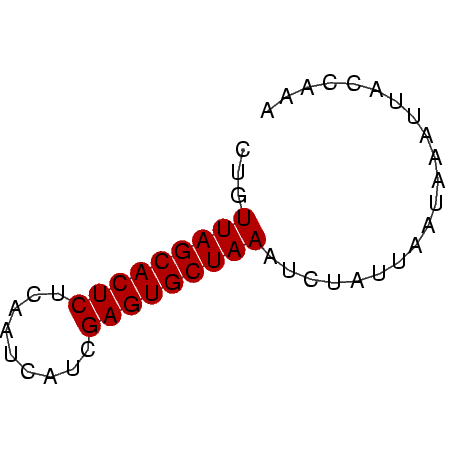
| Location | 4,131,099 – 4,131,150 |
|---|---|
| Length | 51 |
| Sequences | 6 |
| Columns | 51 |
| Reading direction | forward |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -13.77 |
| Consensus MFE | -11.87 |
| Energy contribution | -11.87 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
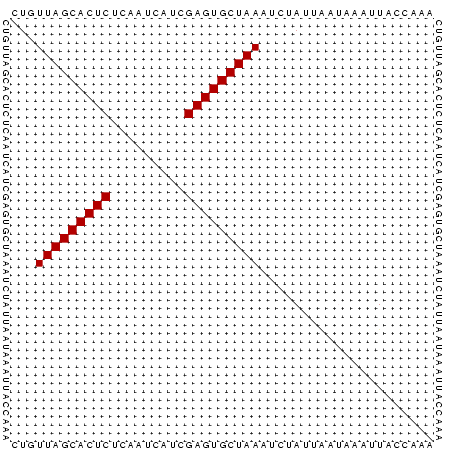
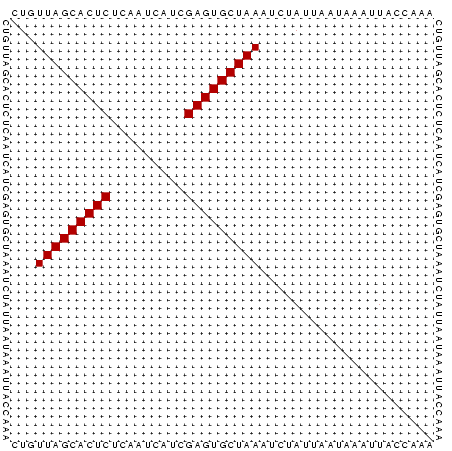
>Banth.0 4131099 51 + 5227293 CUGUUAGCACUCUUUGUCUUCGAGUGCUAAAUCUAUUAAUAACUUACCAAA ...(((((((((.........)))))))))..................... ( -12.00) >Bcere.0 4079729 51 + 5224283 CUGUUAGCACUCUUUGUCUUCGAGUGCUAAAUCUAUUAAUAACUUACCAAA ...(((((((((.........)))))))))..................... ( -12.00) >Bhalo.0 2773051 51 - 4202352 CUGUUAGCACUCUCAUAGUUCGAGUGCUAAAUCUAUUGCUAACUUAUCAAA ...(((((((((.........)))))))))..................... ( -12.00) >Bclau.0 2517408 51 - 4303871 CUGUUAGCACUCAAAUGCAACGAGUGCUAAUACUAUUGAUACACUAUCAAA .(((((((((((.........)))))))))))...((((((...)))))). ( -16.30) >Bsubt.0 2628871 51 + 4214630 CUGUUAGCACUCUCAAUAAGCGAGUGCUAAUUCUAUAACAAAAGUAUCAAA ..((((((((((.(.....).)))))))))).................... ( -13.60) >Blich.0 2636880 51 + 4222334 CUGUUAGCACUCGGCACAACUGAGUGCUAAUUGUAUAUCAAAAUUACCAAA ..((((((((((((.....)))))))))))).................... ( -16.70) >consensus CUGUUAGCACUCUCAAUCAUCGAGUGCUAAAUCUAUUAAUAAAUUACCAAA ...(((((((((.........)))))))))..................... (-11.87 = -11.87 + -0.00)

| Location | 4,131,099 – 4,131,150 |
|---|---|
| Length | 51 |
| Sequences | 6 |
| Columns | 51 |
| Reading direction | reverse |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -13.18 |
| Consensus MFE | -12.25 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
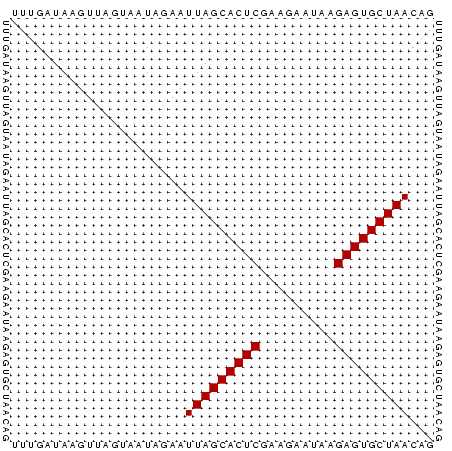
>Banth.0 4131099 51 + 5227293 UUUGGUAAGUUAUUAAUAGAUUUAGCACUCGAAGACAAAGAGUGCUAACAG .....................(((((((((.........)))))))))... ( -12.50) >Bcere.0 4079729 51 + 5224283 UUUGGUAAGUUAUUAAUAGAUUUAGCACUCGAAGACAAAGAGUGCUAACAG .....................(((((((((.........)))))))))... ( -12.50) >Bhalo.0 2773051 51 - 4202352 UUUGAUAAGUUAGCAAUAGAUUUAGCACUCGAACUAUGAGAGUGCUAACAG .....................(((((((((.........)))))))))... ( -12.50) >Bclau.0 2517408 51 - 4303871 UUUGAUAGUGUAUCAAUAGUAUUAGCACUCGUUGCAUUUGAGUGCUAACAG .((((((...)))))).....((((((((((.......))))))))))... ( -15.30) >Bsubt.0 2628871 51 + 4214630 UUUGAUACUUUUGUUAUAGAAUUAGCACUCGCUUAUUGAGAGUGCUAACAG .....................(((((((((.(.....).)))))))))... ( -13.30) >Blich.0 2636880 51 + 4222334 UUUGGUAAUUUUGAUAUACAAUUAGCACUCAGUUGUGCCGAGUGCUAACAG .....................(((((((((.(.....).)))))))))... ( -13.00) >consensus UUUGAUAAGUUAGUAAUAGAAUUAGCACUCGAAGAAUAAGAGUGCUAACAG .....................(((((((((.........)))))))))... (-12.25 = -12.25 + 0.00)

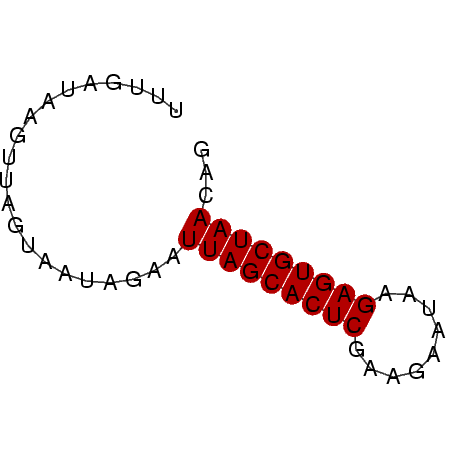

| Location | 4,131,099 – 4,131,150 |
|---|---|
| Length | 51 |
| Sequences | 6 |
| Columns | 51 |
| Reading direction | forward |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -13.77 |
| Consensus MFE | -11.87 |
| Energy contribution | -11.87 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -4.41 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
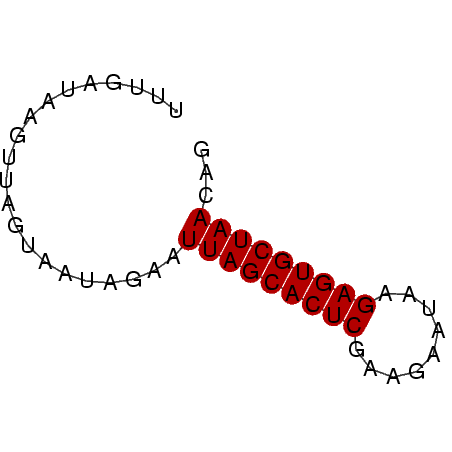
>Banth.0 4131099 51 + 5227293/0-51 CUGUUAGCACUCUUUGUCUUCGAGUGCUAAAUCUAUUAAUAACUUACCAAA ...(((((((((.........)))))))))..................... ( -12.00) >Bcere.0 4079729 51 + 5224283/0-51 CUGUUAGCACUCUUUGUCUUCGAGUGCUAAAUCUAUUAAUAACUUACCAAA ...(((((((((.........)))))))))..................... ( -12.00) >Bhalo.0 2773051 51 - 4202352/0-51 CUGUUAGCACUCUCAUAGUUCGAGUGCUAAAUCUAUUGCUAACUUAUCAAA ...(((((((((.........)))))))))..................... ( -12.00) >Bclau.0 2517408 51 - 4303871/0-51 CUGUUAGCACUCAAAUGCAACGAGUGCUAAUACUAUUGAUACACUAUCAAA .(((((((((((.........)))))))))))...((((((...)))))). ( -16.30) >Bsubt.0 2628871 51 + 4214630/0-51 CUGUUAGCACUCUCAAUAAGCGAGUGCUAAUUCUAUAACAAAAGUAUCAAA ..((((((((((.(.....).)))))))))).................... ( -13.60) >Blich.0 2636880 51 + 4222334/0-51 CUGUUAGCACUCGGCACAACUGAGUGCUAAUUGUAUAUCAAAAUUACCAAA ..((((((((((((.....)))))))))))).................... ( -16.70) >consensus CUGUUAGCACUCUCAAUCAUCGAGUGCUAAAUCUAUUAAUAAAUUACCAAA ...(((((((((.........)))))))))..................... (-11.87 = -11.87 + -0.00)

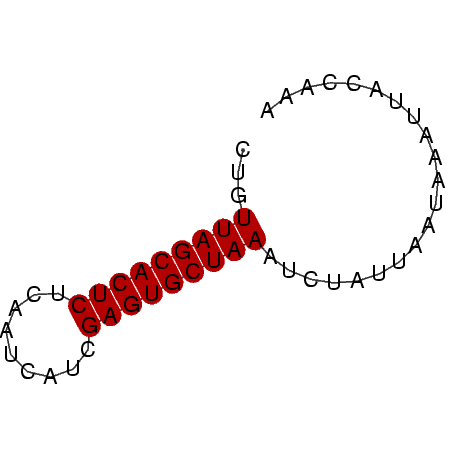
| Location | 4,131,099 – 4,131,150 |
|---|---|
| Length | 51 |
| Sequences | 6 |
| Columns | 51 |
| Reading direction | reverse |
| Mean pairwise identity | 76.34 |
| Mean single sequence MFE | -13.18 |
| Consensus MFE | -12.25 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.89 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.974993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4131099 51 + 5227293/0-51 UUUGGUAAGUUAUUAAUAGAUUUAGCACUCGAAGACAAAGAGUGCUAACAG .....................(((((((((.........)))))))))... ( -12.50) >Bcere.0 4079729 51 + 5224283/0-51 UUUGGUAAGUUAUUAAUAGAUUUAGCACUCGAAGACAAAGAGUGCUAACAG .....................(((((((((.........)))))))))... ( -12.50) >Bhalo.0 2773051 51 - 4202352/0-51 UUUGAUAAGUUAGCAAUAGAUUUAGCACUCGAACUAUGAGAGUGCUAACAG .....................(((((((((.........)))))))))... ( -12.50) >Bclau.0 2517408 51 - 4303871/0-51 UUUGAUAGUGUAUCAAUAGUAUUAGCACUCGUUGCAUUUGAGUGCUAACAG .((((((...)))))).....((((((((((.......))))))))))... ( -15.30) >Bsubt.0 2628871 51 + 4214630/0-51 UUUGAUACUUUUGUUAUAGAAUUAGCACUCGCUUAUUGAGAGUGCUAACAG .....................(((((((((.(.....).)))))))))... ( -13.30) >Blich.0 2636880 51 + 4222334/0-51 UUUGGUAAUUUUGAUAUACAAUUAGCACUCAGUUGUGCCGAGUGCUAACAG .....................(((((((((.(.....).)))))))))... ( -13.00) >consensus UUUGAUAAGUUAGUAAUAGAAUUAGCACUCGAAGAAUAAGAGUGCUAACAG .....................(((((((((.........)))))))))... (-12.25 = -12.25 + 0.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:14:52 2006