
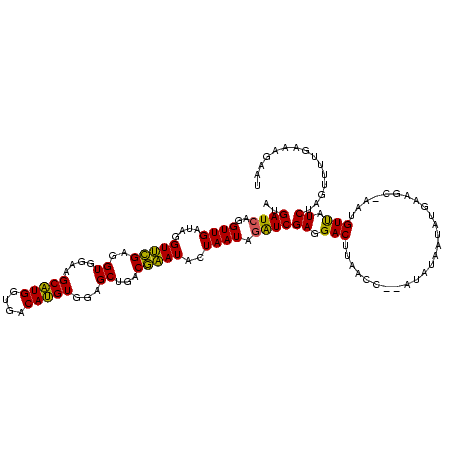
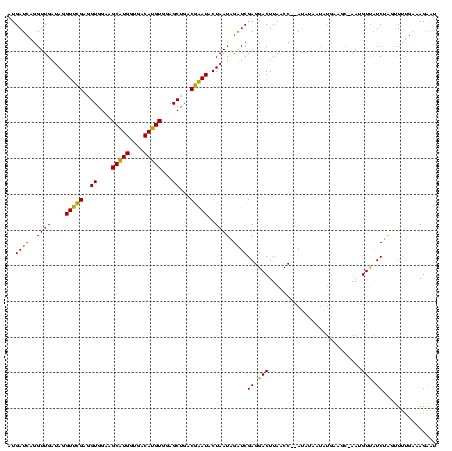
| Sequence ID | Banth.0 |
|---|---|
| Location | 4,652,088 – 4,652,208 |
| Length | 120 |
| Max. P | 0.996860 |

| Location | 4,652,088 – 4,652,205 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Mean single sequence MFE | -29.03 |
| Consensus MFE | -19.47 |
| Energy contribution | -18.04 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.546127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 117 + 5227293/0-120 AUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC--AUAUAAUAUGAAGC-AAUGUUAUCUAGUUUUGAAAGAAU .(((((..((((....(((((.(((....(((((....)))))...)))..)))))..)))).)))))((((((.....--..(((((((.....-.)))))))..))))))........ ( -25.60) >Bcere.0 4651066 118 + 5224283/0-120 AUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC--AUAUAAUAUGAAGCAAAUGUUAUCUAGUUUUGAAAGAAU .(((((..((((....(((((.(((....(((((....)))))...)))..)))))..)))).)))))((((((.....--..(((((((.......)))))))..))))))........ ( -25.10) >Bhalo.0 4174298 119 - 4202352/0-120 AUGAUGAGGUUGAUAGGUCUGGUGUGGAAGCGUGGCGACACGUGAAGCUGACAGAUACUAAUCGGUCGAGGACUUAUCCAAAAACAAAUCAAAAGCAACGUC-UCGAACUCGAGAAGCGU .((((...((((((((((((...((....(((((....)))))...))((((.(((....))).)))).)))))))))....)))..))))...((....((-(((....))))).)).. ( -36.40) >consensus AUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC__AUAUAAUAUGAAGC_AAUGUUAUCUAGUUUUGAAAGAAU ..((((..((((....(((((..((....(((((....)))))...))...)))))..)))).))))((.(((..........................))).))............... (-19.47 = -18.04 + -1.44)

| Location | 4,652,088 – 4,652,206 |
|---|---|
| Length | 118 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.15 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -30.66 |
| Energy contribution | -28.33 |
| Covariance contribution | -2.33 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.763243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 118 + 5227293/40-160 GAGAUUUCCCAUAGCGUAAG-CUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC- ......((((.((((....)-)))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).))).......- ( -35.50) >Bcere.0 4651066 118 + 5224283/40-160 GAGAUUUCCCAUAGCGUAAG-CUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC- ......((((.((((....)-)))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).))).......- ( -35.50) >Bhalo.0 4174298 120 - 4202352/40-160 GAGAUUUCCCAUGGAGUAAAUCCAGUAAGACCCCUUAGAGAUGAUGAGGUUGAUAGGUCUGGUGUGGAAGCGUGGCGACACGUGAAGCUGACAGAUACUAAUCGGUCGAGGACUUAUCCA (.(((.((((.((((.....))))....(((((((((.......)))))..((((((((((..((....(((((....)))))...))...))))).)).)))))))).)))...)))). ( -36.00) >consensus GAGAUUUCCCAUAGCGUAAG_CUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC_ ......((((.((((....).)))....(((((((((.......)))))....((((((((..((....(((((....)))))...))...))))).)))...))))).)))........ (-30.66 = -28.33 + -2.33)

| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -44.77 |
| Consensus MFE | -43.04 |
| Energy contribution | -41.93 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/160-280 AGAGGACCGGGAUGGACGCACCGCUGGUGUACCAGUUGUUCUGCCAAGGGCAUAGCUGGGUAGCUAUGUGCGGAAGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAU .((((.((((.(.(((((....(((((....))))))))))).))..(.((((((((....)))))))).)))..(((....(((((...........)))))....))).))))..... ( -44.20) >Bcere.0 4651066 120 + 5224283/160-280 AGAGGACCGGGAUGGACGCACCGCUGGUGUACCAGUUGUUCUGCCAAGGGCAUAGCUGGGUAGCUAUGUGCGGAAGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAU .((((.((((.(.(((((....(((((....))))))))))).))..(.((((((((....)))))))).)))..(((....(((((...........)))))....))).))))..... ( -44.20) >Bhalo.0 4174298 120 - 4202352/160-280 AGAGGACCGGGAUGGACACACCGCUGGUGUACCAGUUGUUCCGCCAGGAGCAUCGCUGGGUAGCUACGUGUGGACGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAU .((((........(..(((((.(.((((...(((((((((((....))))))..)))))...))))))))))..)(((....(((((...........)))))....))).))))..... ( -45.90) >consensus AGAGGACCGGGAUGGACGCACCGCUGGUGUACCAGUUGUUCUGCCAAGGGCAUAGCUGGGUAGCUAUGUGCGGAAGGGAUAAGUGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAU .((((.((.....)).(((((...((((...(((((((((((....))))))..)))))...)))).)))))...(((....(((((...........)))))....))).))))..... (-43.04 = -41.93 + -1.11)

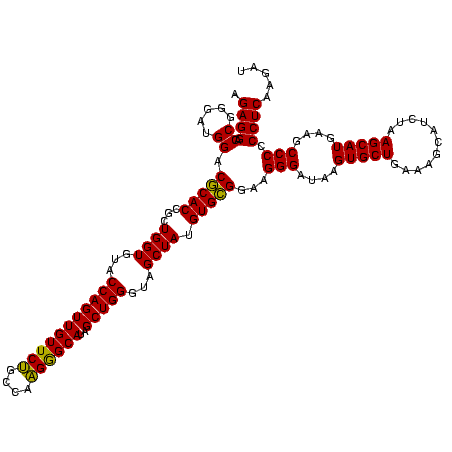
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -53.07 |
| Consensus MFE | -52.40 |
| Energy contribution | -52.73 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.49 |
| SVM RNA-class probability | 0.958440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/320-440 UCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGU ..((((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))((..(.(((((((....))).)))).)..))))))... ( -53.70) >Bcere.0 4651066 120 + 5224283/320-440 UCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGU ..((((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))((..(.(((((((....))).)))).)..))))))... ( -53.70) >Bhalo.0 4174298 120 - 4202352/320-440 UCGAUGUCGGCUCAUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGU ..((((.((((((((((((((((((((((......)))))).)))))(((((.....))))).....)))))....))))))((..(.(((((((....))).)))).)..))))))... ( -51.80) >consensus UCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGU ..((((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))((..(.(((((((....))).)))).)..))))))... (-52.40 = -52.73 + 0.33)

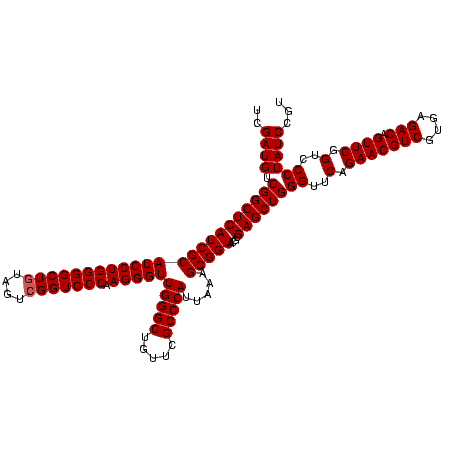
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -50.83 |
| Consensus MFE | -50.29 |
| Energy contribution | -50.40 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608048 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/360-480 UCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGC .(((......(((((.(((((((.((............))))))))).)))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))). ( -51.90) >Bcere.0 4651066 120 + 5224283/360-480 UCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGC .(((......(((((.(((((((.((............))))))))).)))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))). ( -51.90) >Bhalo.0 4174298 120 - 4202352/360-480 UCUCCCCCAAGAGUUCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGAAGUAGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGC ............((((....(((((..(((((.....)))))..)))))((..((((((((((((((((......)))))).)))))(((((.....))))).....)))))..)))))) ( -48.70) >consensus UCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGC .(((......(((((.(((((((.((............))))))))).)))))(((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))). (-50.29 = -50.40 + 0.11)

| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -44.93 |
| Consensus MFE | -44.93 |
| Energy contribution | -44.27 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.91 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/480-600 AGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUA ((((((.((((..(((......)))..)))).(((....)))(((..((((.((((((((....)))......((((....))))..)))))....)))).)))........)))))).. ( -45.20) >Bcere.0 4651066 120 + 5224283/480-600 AGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUA ((((((.((((..(((......)))..)))).(((....)))(((..((((.((((((((....)))......((((....))))..)))))....)))).)))........)))))).. ( -45.20) >Bhalo.0 4174298 120 - 4202352/480-600 AGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGCGGUGCCGUAUGGAAGGGCCGUCGCUCAACGGAUAAAAGCUACCCUGGGGAUAACAGGCUUA ((((((.((((..(((......)))..)))).(((....)))(((..((((.((((((((((.((........))))))))(.....)))))....))))))).........)))))).. ( -44.40) >consensus AGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUA ((((((.((((..(((......)))..)))).(((....)))(((..((((.((((((((....)))......((((....))))..)))))....)))).)))........)))))).. (-44.93 = -44.27 + -0.66)

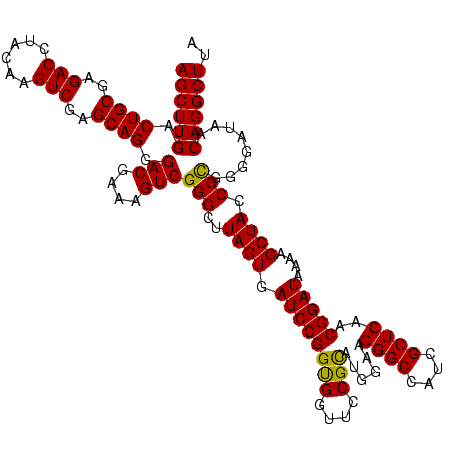
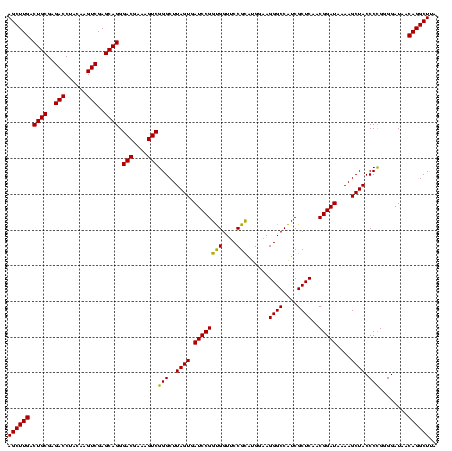
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -39.60 |
| Consensus MFE | -39.09 |
| Energy contribution | -38.77 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.814932 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/520-640 AAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAU .((((((.....(((((......((((....))))....((((((..((((..(((......)))..)))).(((....))))))))))))))(((((((....))).))))..)))))) ( -39.90) >Bcere.0 4651066 120 + 5224283/520-640 AAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAU .((((((.....(((((......((((....))))....((((((..((((..(((......)))..)))).(((....))))))))))))))(((((((....))).))))..)))))) ( -39.90) >Bhalo.0 4174298 120 - 4202352/520-640 AAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAAAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGCGGUGCCGUAUGGAAGGGCCGU ....((((((..(((((.......(((....))).....((((((..((((..(((......)))..)))).(((....))))))))))))))))))))(((.((........))))).. ( -39.00) >consensus AAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAU .((((((.....(((((......((((....))))....((((((..((((..(((......)))..)))).(((....))))))))))))))(((((((....))).))))..)))))) (-39.09 = -38.77 + -0.33)

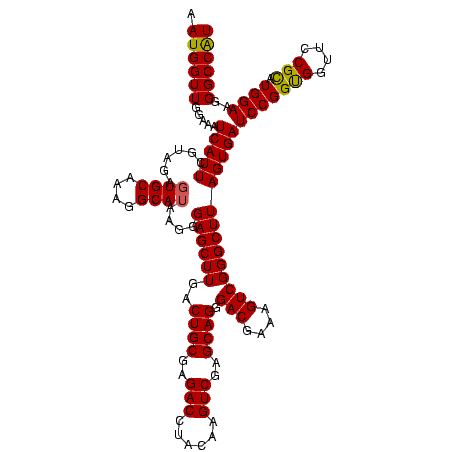
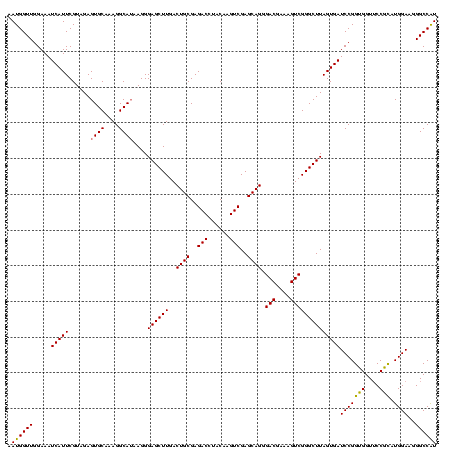
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -35.10 |
| Consensus MFE | -34.67 |
| Energy contribution | -35.00 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697914 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/560-680 CUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGG ....(((((((((..((((....))))..))))......((((((((((....)))......(((((.........))))))))))))......)))))(((((...........))))) ( -35.60) >Bcere.0 4651066 120 + 5224283/560-680 CUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGG ....(((((((((..((((....))))..))))......((((((((((....)))......(((((.........))))))))))))......)))))(((((...........))))) ( -35.60) >Bhalo.0 4174298 120 - 4202352/560-680 CUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUUUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACUGUUUAGGAGG ....(((((((((..((((....))))..))))......((((((.(((....)))......(((((.........))))).))))))......)))))(((((...........))))) ( -34.10) >consensus CUUUCGUCCCUGCUCGACUUGUAGGUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGG ....(((((((((..((((....))))..))))......((((((((((....)))......(((((.........))))))))))))......)))))(((((...........))))) (-34.67 = -35.00 + 0.33)

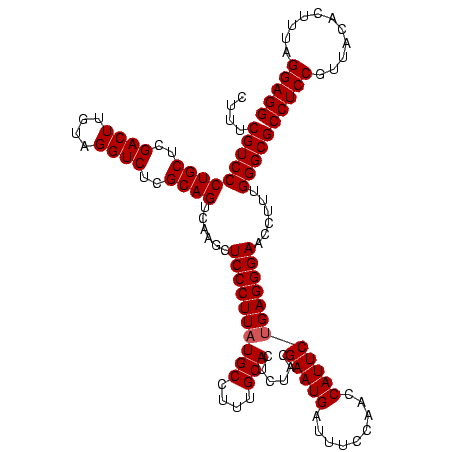
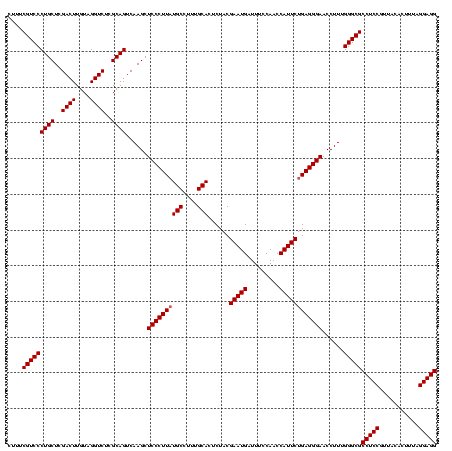
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.73 |
| Consensus MFE | -41.07 |
| Energy contribution | -41.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.90 |
| SVM RNA-class probability | 0.982010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/560-680 CCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAG (((((...........)))))(((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))...(((..(((......)))..)))))).)).... ( -42.20) >Bcere.0 4651066 120 + 5224283/560-680 CCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAG (((((...........)))))(((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))...(((..(((......)))..)))))).)).... ( -42.20) >Bhalo.0 4174298 120 - 4202352/560-680 CCUCCUAAACAGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAAAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAG (((((...........)))))(((((..(((((((((..(((((((.....)))))))......(((....)))..)))))))))...(((..(((......)))..)))))).)).... ( -40.80) >consensus CCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAG (((((...........)))))(((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))...(((..(((......)))..)))))).)).... (-41.07 = -41.40 + 0.33)

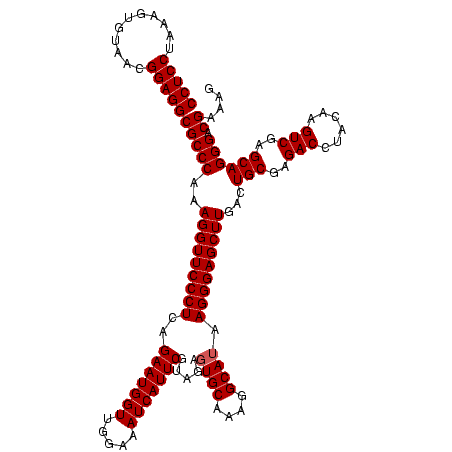
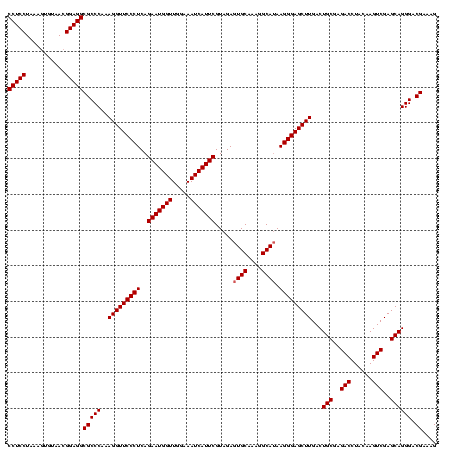
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -34.57 |
| Consensus MFE | -34.07 |
| Energy contribution | -34.40 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964767 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/600-720 CCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCC (((((((((....)))......(((((.........)))))))))))......(((((((((((...........))))))(((.......))).....)))))...(((...))).... ( -35.10) >Bcere.0 4651066 120 + 5224283/600-720 CCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCC (((((((((....)))......(((((.........)))))))))))......(((((((((((...........))))))(((.......))).....)))))...(((...))).... ( -35.10) >Bhalo.0 4174298 120 - 4202352/600-720 CCCUUUUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCCCUG (((((.(((....)))......(((((.........))))).)))))......(((((((((((...........))))))(((.......))).....)))))................ ( -33.50) >consensus CCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCC (((((((((....)))......(((((.........)))))))))))......(((((((((((...........))))))(((.......))).....)))))................ (-34.07 = -34.40 + 0.33)

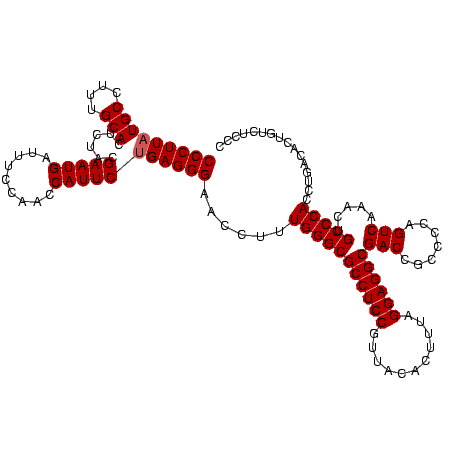
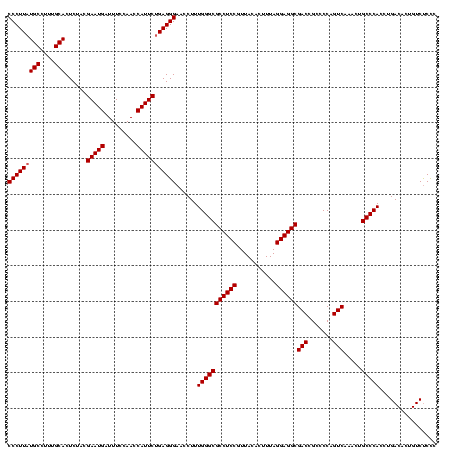
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -44.03 |
| Consensus MFE | -43.06 |
| Energy contribution | -40.73 |
| Covariance contribution | -2.33 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991146 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/640-760 CUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCA ((((..(((....(((((((((((...........))))))(((.......))).....))))).(((((.(((.....((((((.....))))))))))))))...)))..).)))... ( -42.00) >Bcere.0 4651066 120 + 5224283/640-760 CUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCA ((((..(((....(((((((((((...........))))))(((.......))).....))))).(((((.(((.....((((((.....))))))))))))))...)))..).)))... ( -42.00) >Bhalo.0 4174298 120 - 4202352/640-760 CUGAGGGAACCUUUGGGCGCCUCCGUUACUGUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCCCUGAACCGGAUCACGGUUCGAGGUUAGAAUGUCAAUACCGUCA ..((.((......(((((((((((...........))))))(((.......))).....)))))..((((((((.((.(((((((.....))))))).)).)))..)))))...)).)). ( -48.10) >consensus CUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCA .(((((.(((((.(((((((((((...........))))))(((.......))).....)))))...............((((((.....)))))).)))))....)))))......... (-43.06 = -40.73 + -2.33)

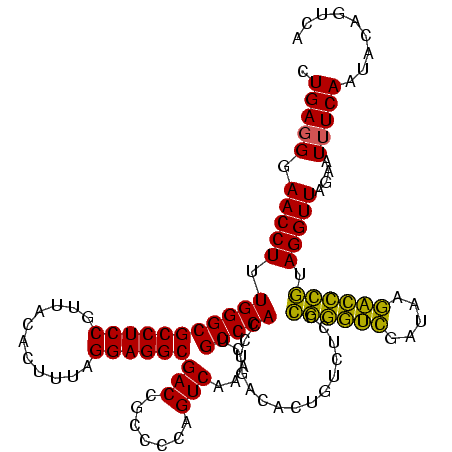
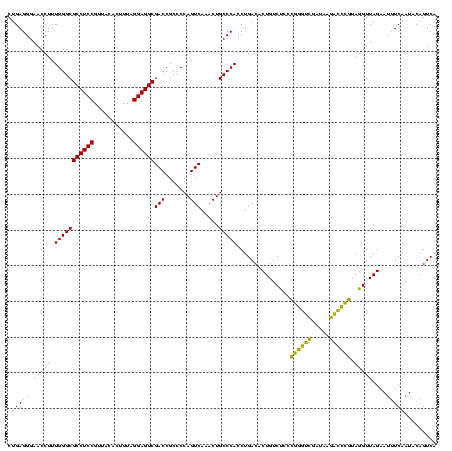
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -44.90 |
| Consensus MFE | -42.73 |
| Energy contribution | -40.07 |
| Covariance contribution | -2.66 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.707172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/640-760 UGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAG ...(((....(((......((((.((((((.....))))))(....)......))))((((.....(((((...)))))((((((...........)))))))))).....)))...))) ( -43.10) >Bcere.0 4651066 120 + 5224283/640-760 UGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAG ...(((....(((......((((.((((((.....))))))(....)......))))((((.....(((((...)))))((((((...........)))))))))).....)))...))) ( -43.10) >Bhalo.0 4174298 120 - 4202352/640-760 UGACGGUAUUGACAUUCUAACCUCGAACCGUGAUCCGGUUCAGGGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAACAGUAACGGAGGCGCCCAAAGGUUCCCUCAG (((.((..((((((((....(((.((((((.....)))))))))...)))))))).(((((.....(((((...)))))((((((...........)))))))))))......)).))). ( -48.50) >consensus UGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAG ..................(((((.((((((.....))))))...(((...)))...(((((.....(((((...)))))((((((...........))))))))))).)))))....... (-42.73 = -40.07 + -2.66)

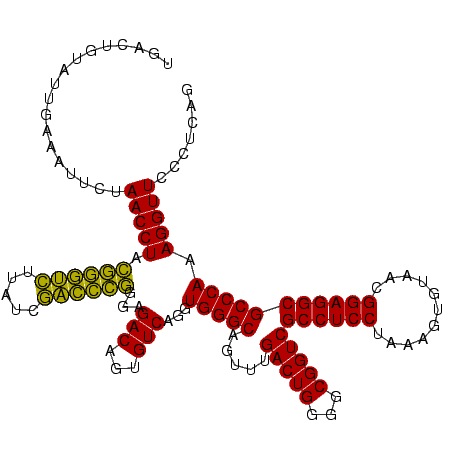
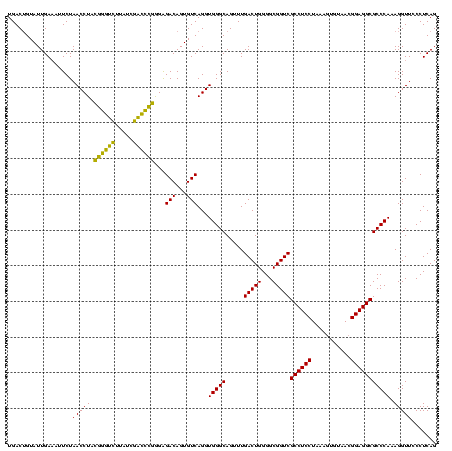
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -37.24 |
| Consensus MFE | -38.61 |
| Energy contribution | -35.84 |
| Covariance contribution | -2.77 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.57 |
| Structure conservation index | 1.04 |
| SVM decision value | 1.86 |
| SVM RNA-class probability | 0.980172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/680-800 CGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUC ..(((((((..((((...........)))).(((.((..((((((.....))))))..)).)))................)))))))........(((((..............))))). ( -38.24) >Bcere.0 4651066 120 + 5224283/680-800 CGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUC ..(((((((..((((...........)))).(((.((..((((((.....))))))..)).)))................)))))))........(((((..............))))). ( -38.24) >Bhalo.0 4174298 120 - 4202352/680-800 CGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCCCUGAACCGGAUCACGGUUCGAGGUUAGAAUGUCAAUACCGUCAGGGUAGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUC .(((.......)))..(((((((((.((((((((.((.(((((((.....))))))).)).)))..))))).....))..)))))))........(((((..............))))). ( -35.24) >consensus CGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUC ..(((((((..((((...........)))).(((.((..((((((.....))))))..)).)))................)))))))........(((((..............))))). (-38.61 = -35.84 + -2.77)

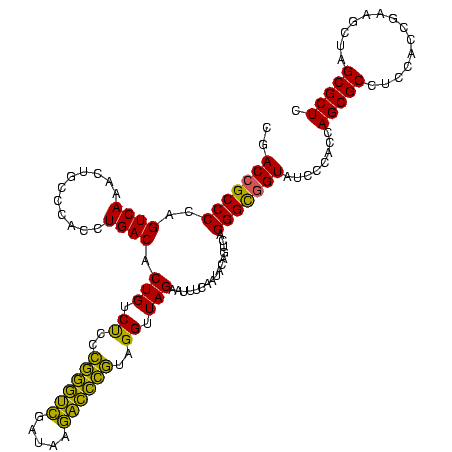
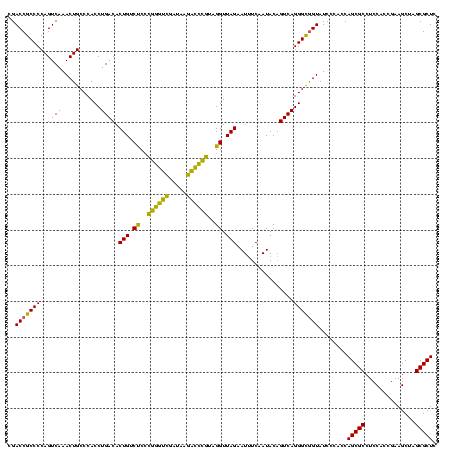
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.89 |
| Mean single sequence MFE | -48.63 |
| Consensus MFE | -48.89 |
| Energy contribution | -45.57 |
| Covariance contribution | -3.32 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.27 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.880745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/680-800 GAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCG ((.((((.(((((....))))).(..((.((((.(((((.((((....................((((((.....))))))(....)...)))))))))...)))).))..))))).)). ( -49.40) >Bcere.0 4651066 120 + 5224283/680-800 GAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCG ((.((((.(((((....))))).(..((.((((.(((((.((((....................((((((.....))))))(....)...)))))))))...)))).))..))))).)). ( -49.40) >Bhalo.0 4174298 120 - 4202352/680-800 GAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACUACCCUGACGGUAUUGACAUUCUAACCUCGAACCGUGAUCCGGUUCAGGGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCG .((((((.((....))...))))))(((((....)))))..(((.((.((((((((....(((.((((((.....)))))))))...)))))))).....(((.....)))..)).))). ( -47.10) >consensus GAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCG ((.((((.(((((....))))).(..((.((((.(((((.((((...((((....(((......((((((.....))))))..))))))))))))))))...)))).))..))))).)). (-48.89 = -45.57 + -3.32)

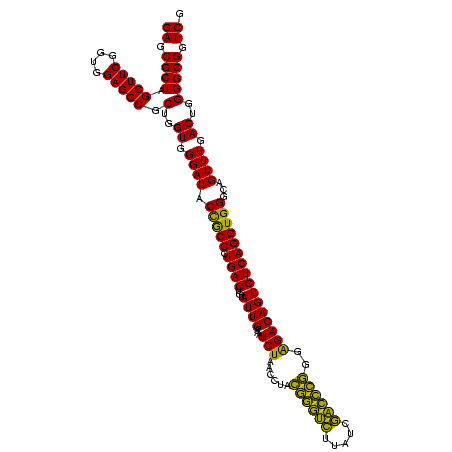
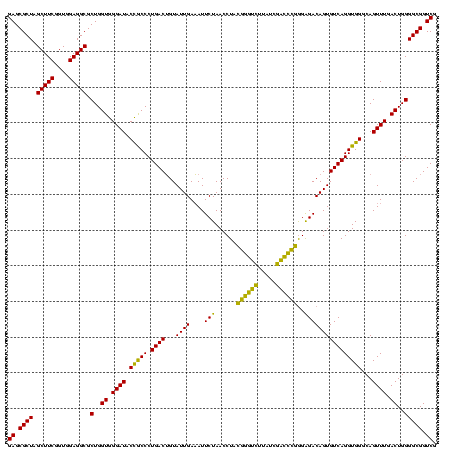
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -35.41 |
| Energy contribution | -32.20 |
| Covariance contribution | -3.21 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.95 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.60 |
| SVM RNA-class probability | 0.794638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
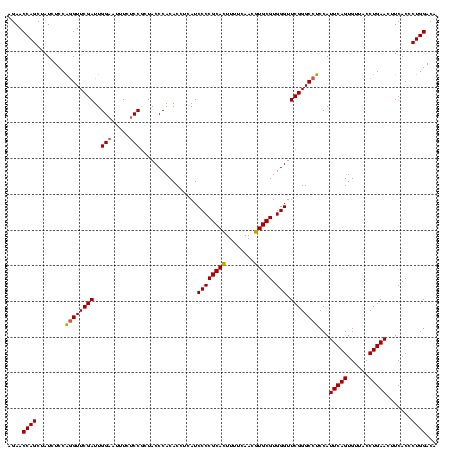
>Banth.0 4652088 120 + 5227293/720-840 GGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGUACC (((((.....)))))(((((((.(.....)..(((((...(((((((.....)))...((((..((((.(((....))).)))).....))))....))))...)))))..)))).))). ( -37.40) >Bcere.0 4651066 120 + 5224283/720-840 GGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGUACC (((((.....)))))(((((((.(.....)..(((((...(((((((.....)))...((((..((((.(((....))).)))).....))))....))))...)))))..)))).))). ( -37.40) >Bhalo.0 4174298 120 - 4202352/720-840 AACCGGAUCACGGUUCGAGGUUAGAAUGUCAAUACCGUCAGGGUAGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGCUUCCAAGGCUCCUACCUAUCCUGUACAAACGGUACC (((((.....))))).((.((....)).))..((((((((((((((.....(....).((((...(((.(((....))).)))......))))......)))))))).....)))))).. ( -31.60) >consensus GGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGUACC (((((.....))))).................((((((((((((((..........(.((((..((((.(((....))).)))).....)))).)))))....)))).....)))))).. (-35.41 = -32.20 + -3.21)

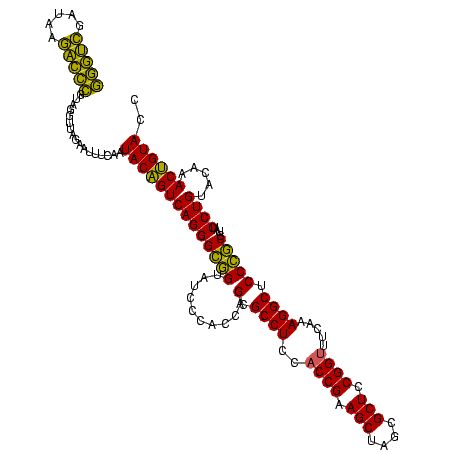
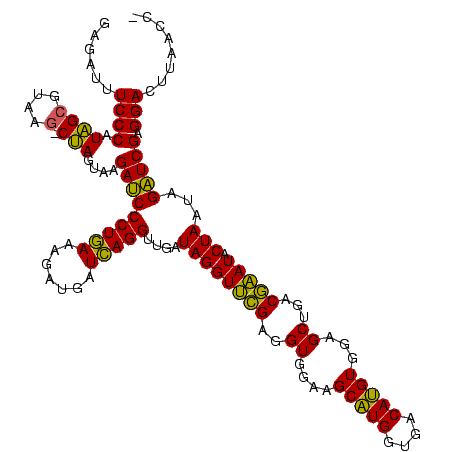
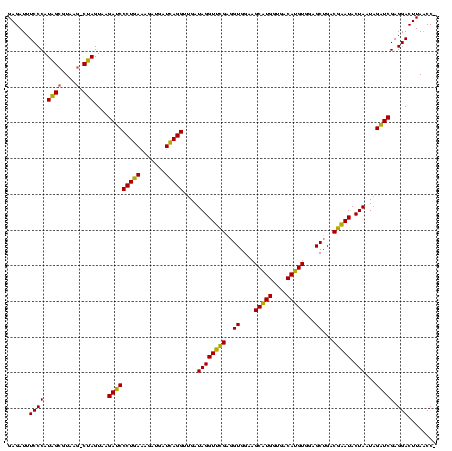
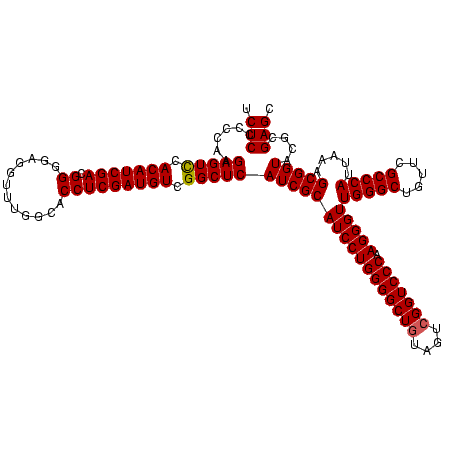
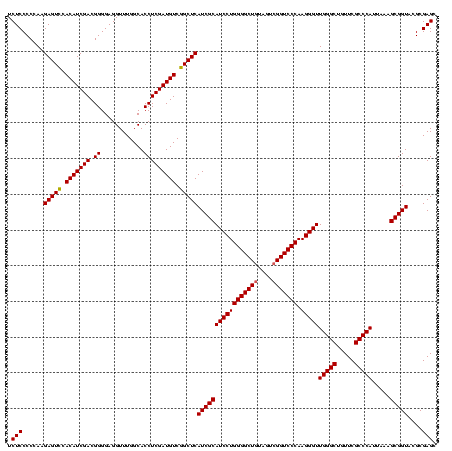
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.78 |
| Mean single sequence MFE | -46.80 |
| Consensus MFE | -49.59 |
| Energy contribution | -45.83 |
| Covariance contribution | -3.76 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.78 |
| Structure conservation index | 1.06 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/720-840 GGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCC (((..(((((((((((..(((((((..(((((....((((((((....)))))))).))))).)))((((....)))))))).))))))..)))))...)))...(((((.....))))) ( -48.40) >Bcere.0 4651066 120 + 5224283/720-840 GGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCC (((..(((((((((((..(((((((..(((((....((((((((....)))))))).))))).)))((((....)))))))).))))))..)))))...)))...(((((.....))))) ( -48.40) >Bhalo.0 4174298 120 - 4202352/720-840 GGUACCGUUUGUACAGGAUAGGUAGGAGCCUUGGAAGCCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACUACCCUGACGGUAUUGACAUUCUAACCUCGAACCGUGAUCCGGUU .((((.....)))).((((.((((((.(((((....((((((((....)))))))).)))))((((..(((......)))...)))).............)))...)))...)))).... ( -43.60) >consensus GGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCC ((((((((.....((((.....(((..(((((....((((((((....)))))))).))))).)))((((....))))))))))))))))...............(((((.....))))) (-49.59 = -45.83 + -3.76)


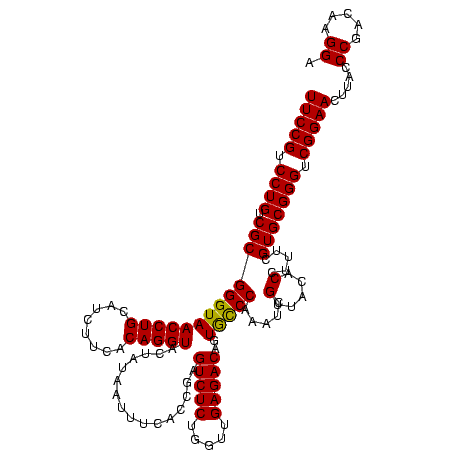
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.60 |
| Consensus MFE | -36.71 |
| Energy contribution | -36.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575833 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/880-1000 UUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUGGUUGAGACAGUGCCCAAAUCGUUACACCUUUCGUGCGGGUCGGAACUUACCCGACAAGGA (((((.((((.(((((((((((((........)))))................(((((.....)))))..))))).....(.....).....))))))).))))).....((.....)). ( -37.20) >Bcere.0 4651066 120 + 5224283/880-1000 UUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUGGUUGAGACAGUGCCCAAAUCGUUACACCUUUCGUGCGGGUCGGAACUUACCCGACAAGGA (((((.((((.(((((((((((((........)))))................(((((.....)))))..))))).....(.....).....))))))).))))).....((.....)). ( -37.20) >Bhalo.0 4174298 120 - 4202352/880-1000 UUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGGGUCUCUCGUUGAGACAGUAUCCAAGUCGUUACACCAUUCGUGCGGGUCGGAACUUACCCGACAAGGA (((((.((((.(((((((.(((((........)))))..............(((((((.....))))).(((..........))).))))))))))))).))))).....((.....)). ( -38.40) >consensus UUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUGGUUGAGACAGUGCCCAAAUCGUUACACCUUUCGUGCGGGUCGGAACUUACCCGACAAGGA (((((.((((.(((((((((((((........)))))................(((((.....)))))..))))).....(.....).....))))))).))))).....((.....)). (-36.71 = -36.27 + -0.44)

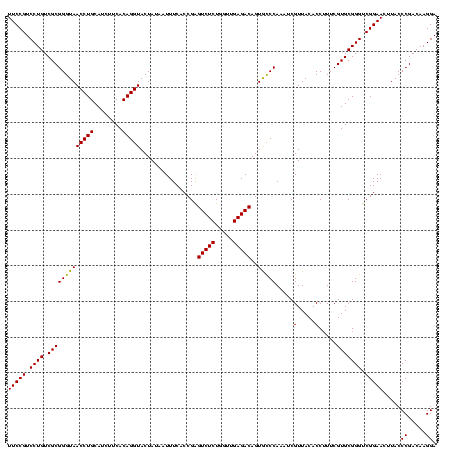
| Location | 4,652,088 – 4,652,205 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.72 |
| Mean single sequence MFE | -42.93 |
| Consensus MFE | -38.12 |
| Energy contribution | -37.23 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 117 + 5227293/1000-1120 GGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC---GUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAU ..(....)(((((((...((((((..((((..((...((((((.((---....))..))))))...))..))))))))))...)))((((((.......))))))...))))........ ( -42.30) >Bcere.0 4651066 117 + 5224283/1000-1120 GGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC---GUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAU ..(....)(((((((...((((((..((((..((...((((((.((---....))..))))))...))..))))))))))...)))((((((.......))))))...))))........ ( -42.30) >Bhalo.0 4174298 120 - 4202352/1000-1120 GGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAGGGGUUAUCCCUUCGGGAGAAGCUCUGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAU ..(((...(((((((...((((((..((((..((...((((((.((((....)).))))))))...))..))))))))))...)))((((((.......))))))...)))))))..... ( -44.20) >consensus GGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC___GUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAU ....((..(((((((...((((((..((((..((...((((((.((.......))..))))))...))..))))))))))...)))((((((.......))))))...))))..)).... (-38.12 = -37.23 + -0.89)

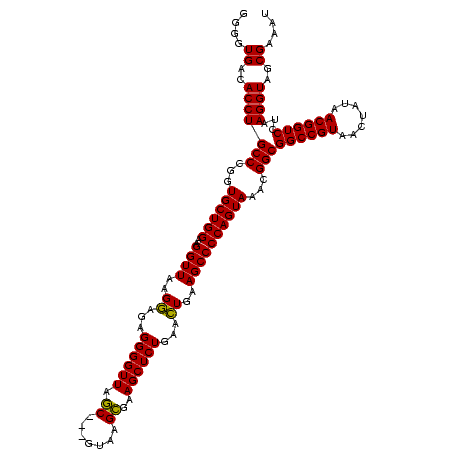
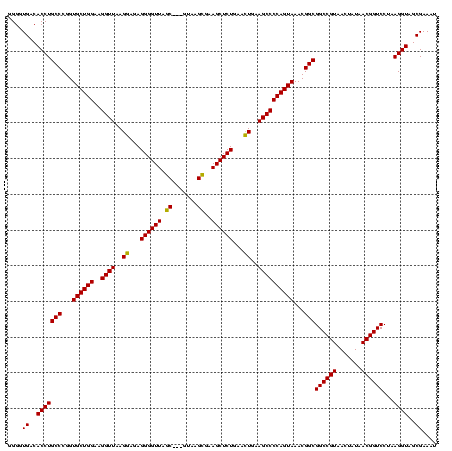
| Location | 4,652,088 – 4,652,205 |
|---|---|
| Length | 117 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.60 |
| Mean single sequence MFE | -42.37 |
| Consensus MFE | -37.93 |
| Energy contribution | -36.60 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 117 + 5227293/1040-1160 AAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC---GUAAGCGAAGCUCUGAACUGAAGCCCCAGUA .....(((..((((.(((...(((....)))...))).)))).))).((((.....))))((((..((((..((...((((((.((---....))..))))))...))..)))))))).. ( -42.10) >Bcere.0 4651066 117 + 5224283/1040-1160 AAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC---GUAAGCGAAGCUCUGAACUGAAGCCCCAGUA .....(((..((((.(((...(((....)))...))).)))).))).((((.....))))((((..((((..((...((((((.((---....))..))))))...))..)))))))).. ( -42.10) >Bhalo.0 4174298 120 - 4202352/1040-1160 AAAAACACAGGUCUCUGCGAAGCCGCAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAGGAGGGGUUAUCCCUUCGGGAGAAGCUCUGAAUUGAAGCCCCAGUA ............((.(((...(((....)))...))).))((((........))))..((((((..((((..((...((((((.((((....)).))))))))...))..)))))))))) ( -42.90) >consensus AAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGC___GUAAGCGAAGCUCUGAACUGAAGCCCCAGUA .........(((((((((...(((....)))...))..)))))))..((((.....))))((((..((((..((...((((((.((.......))..))))))...))..)))))))).. (-37.93 = -36.60 + -1.33)

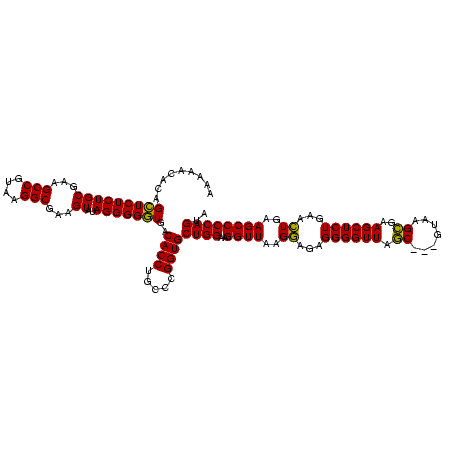
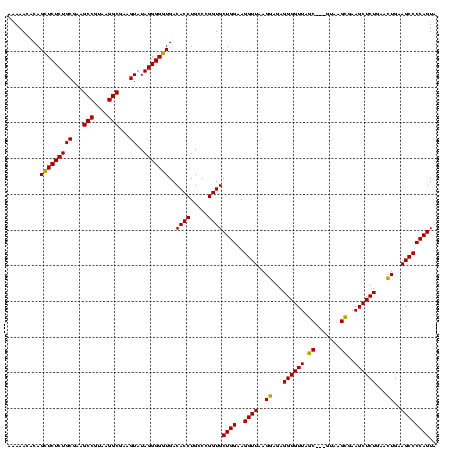
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -23.49 |
| Energy contribution | -22.16 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.66 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/1400-1520 CCAUCACAGCUCAGCCUUCACGAUAAGCGGAUUUGCCUACUUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGA .......(((...........((((((..(......)..))))))(((......))).((((((((..........))))((...............)))))).......)))....... ( -21.16) >Bcere.0 4651066 120 + 5224283/1400-1520 CCAUCACAGCUCAGCCUUCACGAUAAGCGGAUUUGCCUACUUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGA .......(((...........((((((..(......)..))))))(((......))).((((((((..........))))((...............)))))).......)))....... ( -21.16) >Bhalo.0 4174298 120 - 4202352/1400-1520 CCAUCACAGCUCAGCCUUUAUGGAAAGCGGAUUUGCCUACUUUCCAGCCUAACUGCUUGGACGCGCAUCUCCAUCAGCGCGCUUACCCUACCUUUCUGCGUCCCCCCAUUGCUCAAACGG ((.....(((.(((......(((((((..(......)..)))))))......)))...((((((((..........))))((...............)))))).......))).....)) ( -26.76) >consensus CCAUCACAGCUCAGCCUUCACGAUAAGCGGAUUUGCCUACUUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGA .......(((...........((((((..(......)..))))))(((......))).((((((((..........))))((...............)))))).......)))....... (-23.49 = -22.16 + -1.33)

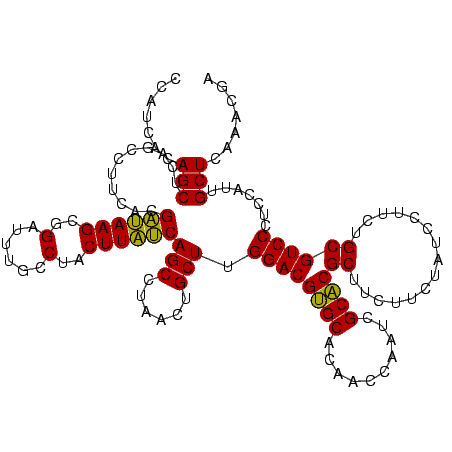
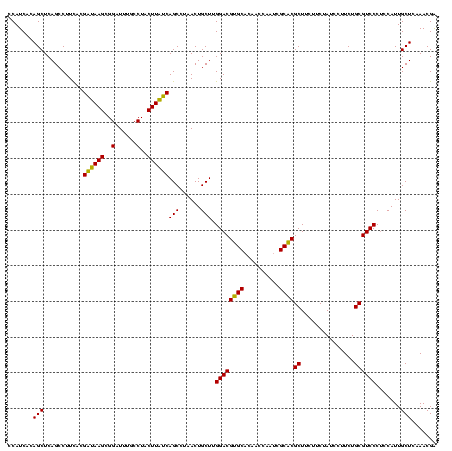
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.33 |
| Mean single sequence MFE | -30.81 |
| Consensus MFE | -29.77 |
| Energy contribution | -28.99 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821187 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/1440-1560 UUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGC ((((((((......))).((((((((..........))))((...............))))))...............)))))...(((((.(((((........)))))...))))).. ( -28.86) >Bcere.0 4651066 120 + 5224283/1440-1560 UUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGC ((((((((......))).((((((((..........))))((...............))))))...............)))))...(((((.(((((........)))))...))))).. ( -28.86) >Bhalo.0 4174298 120 - 4202352/1440-1560 UUUCCAGCCUAACUGCUUGGACGCGCAUCUCCAUCAGCGCGCUUACCCUACCUUUCUGCGUCCCCCCAUUGCUCAAACGGUGAGGAGGUGGUACAGGAAUUUCCACCUGUUGUCCAUCGC ..((((((......)).))))(((((..........)))))....((.(((((((..(((.........)))..))).)))).)).(((((.(((((........)))))...))))).. ( -34.70) >consensus UUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGC ((((((((......))).((((((((..........))))((...............))))))...............)))))...(((((.(((((........)))))...))))).. (-29.77 = -28.99 + -0.77)


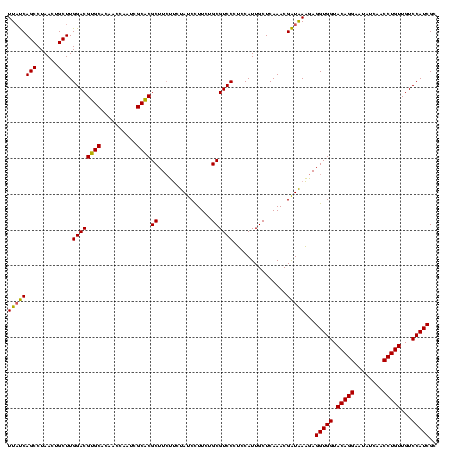
| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.99 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -38.70 |
| Energy contribution | -38.03 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.93 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511781 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/1560-1680 CUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUU-CGCUACUCAUACCGGCA ....(((((((((((((.(((((((...........))))))).)).).)))((((...))))......))))..(((.(((((((.......))))))).)-))...........))). ( -41.50) >Bcere.0 4651066 119 + 5224283/1560-1680 CUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUU-CGCUACUCAUACCGGCA ....(((((((((((((.(((((((...........))))))).)).).)))((((...))))......))))..(((.(((((((.......))))))).)-))...........))). ( -41.50) >Bhalo.0 4174298 120 - 4202352/1560-1680 CUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUGGGCUUUCGACGGAGGGGAUUCUCACCCCUCUUUUCGCUACUCAUACCGGCA ....(((....((((((.(((((((...........))))))).))((((..((((...))))..))))))))..(((.(((((((.......)))))))..)))...........))). ( -41.50) >consensus CUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUU_CGCUACUCAUACCGGCA ....(((....((((((.(((((((...........))))))).)).).)))..(((.(((...))).)))....((..(((((((.......)))))))...))...........))). (-38.70 = -38.03 + -0.66)

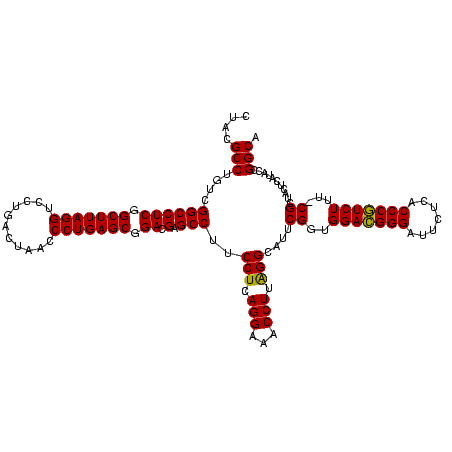
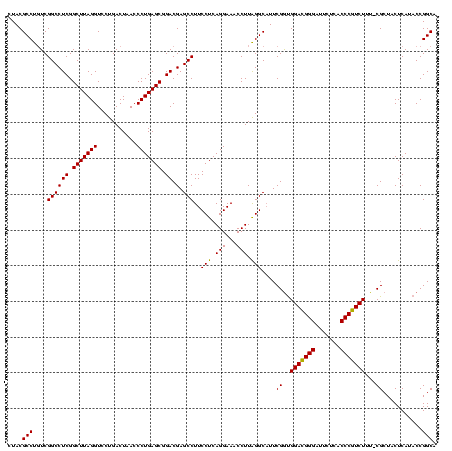
| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.09 |
| Mean single sequence MFE | -24.49 |
| Consensus MFE | -25.75 |
| Energy contribution | -24.42 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.17 |
| Structure conservation index | 1.05 |
| SVM decision value | 2.55 |
| SVM RNA-class probability | 0.995167 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/1640-1760 GACGGGAUUCUCACCCGUCUUU-CGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU ((((((.......))))))...-.(((.........)))........(((((((.((.....(((.((....)).)))........)).)))))))........................ ( -22.72) >Bcere.0 4651066 119 + 5224283/1640-1760 GACGGGAUUCUCACCCGUCUUU-CGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU ((((((.......))))))...-.(((.........)))........(((((((.((.....(((.((....)).)))........)).)))))))........................ ( -22.72) >Bhalo.0 4174298 120 - 4202352/1640-1760 GAGGGGAUUCUCACCCCUCUUUUCGCUACUCAUACCGGCAUUCUCACUUCUACGCGCUCCACCAGUCCUCUCGGUCUGACUUCGCUGCACGUAGAACGCUCCCCUACCACUCGCACCAAC ((((((.......)))))).................((.........(((((((.((.....(((.((....)).)))........)).))))))).((.............)).))... ( -28.04) >consensus GACGGGAUUCUCACCCGUCUUU_CGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU ((((((.......)))))).....(((.........)))........(((((((.((.....(((.((....)).)))........)).)))))))........................ (-25.75 = -24.42 + -1.33)

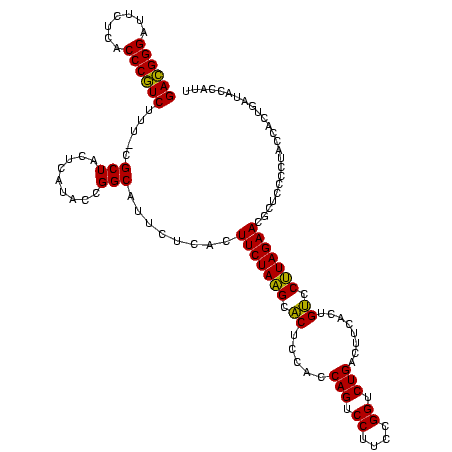
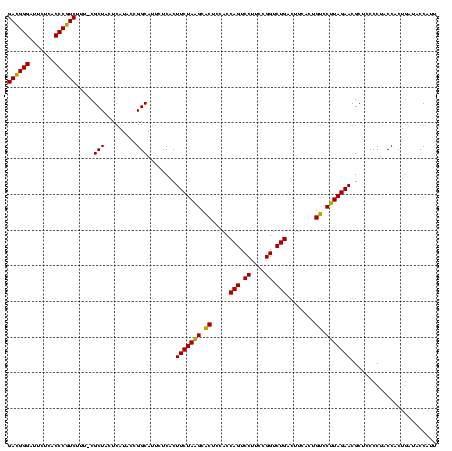
| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.09 |
| Mean single sequence MFE | -38.93 |
| Consensus MFE | -37.10 |
| Energy contribution | -35.33 |
| Covariance contribution | -1.77 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988727 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/1640-1760 AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG-AAAGACGGGUGAGAAUCCCGUC ..((.(((((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))...))).)).))-...((((((.......)))))) ( -35.70) >Bcere.0 4651066 119 + 5224283/1640-1760 AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG-AAAGACGGGUGAGAAUCCCGUC ..((.(((((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))...))).)).))-...((((((.......)))))) ( -35.70) >Bhalo.0 4174298 120 - 4202352/1640-1760 GUUGGUGCGAGUGGUAGGGGAGCGUUCUACGUGCAGCGAAGUCAGACCGAGAGGACUGGUGGAGCGCGUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAAGAGGGGUGAGAAUCCCCUC .(((.(((.(.(((((.....((.((((((((((..((...((((.((....)).))))))..)))))))))))).....))))).)....))).)))...((((((.......)))))) ( -45.40) >consensus AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCG_AAAGACGGGUGAGAAUCCCGUC ...(.(((.(.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).)....))).).....((((((.......)))))) (-37.10 = -35.33 + -1.77)

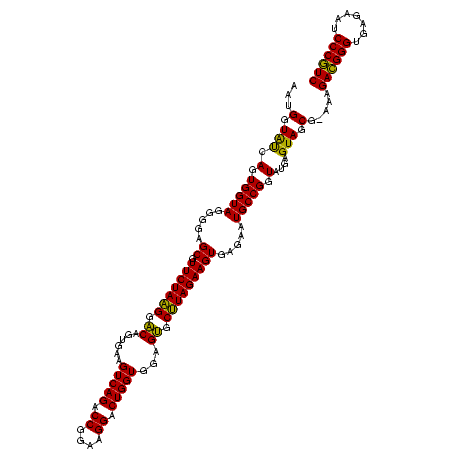
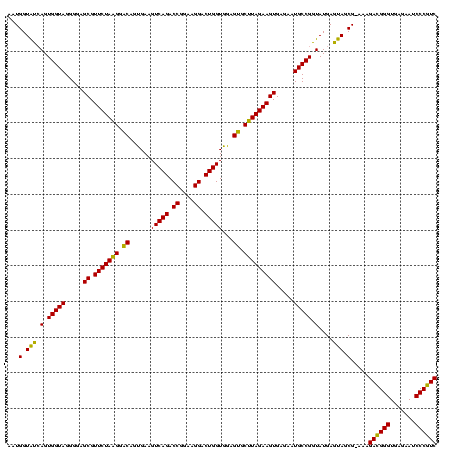
| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -30.09 |
| Energy contribution | -27.99 |
| Covariance contribution | -2.10 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.72 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.990021 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/1680-1800 UUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU-GGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGG .........(((((.((.(((((((.((....)).))(((......)))....(((.(((..........(((((((...-)))))))......))))))))))).)).)))))...... ( -27.79) >Bcere.0 4651066 120 + 5224283/1680-1800 UUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUCGGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGG .........(((((.((.(((((((.((....)).))(((......)))....(((.(((..........(((((((....)))))))......))))))))))).)).)))))...... ( -29.39) >Bhalo.0 4174298 119 - 4202352/1680-1800 UUCUCACUUCUACGCGCUCCACCAGUCCUCUCGGUCUGACUUCGCUGCACGUAGAACGCUCCCCUACCACUCGCACCAAC-GGUGCGAAUCCAUAGCUUCGGUGAUACGUUUAGCCCCGG .......(((((((.((.....(((.((....)).)))........)).)))))))..........((..(((((((...-))))))).......(((.((......))...)))...)) ( -31.22) >consensus UUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU_GGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGG .......(((((((.((.....(((.((....)).)))........)).))))))).(((....(((((((((((((....)))))))...(........))))))).....)))..... (-30.09 = -27.99 + -2.10)

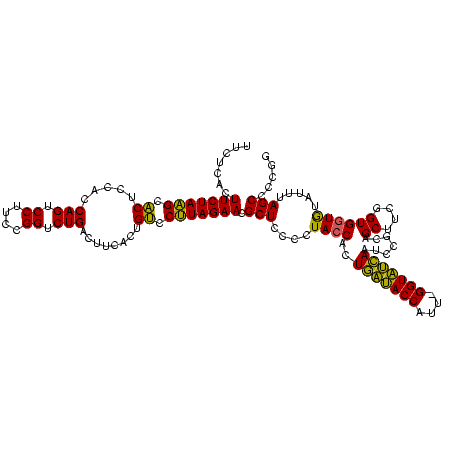
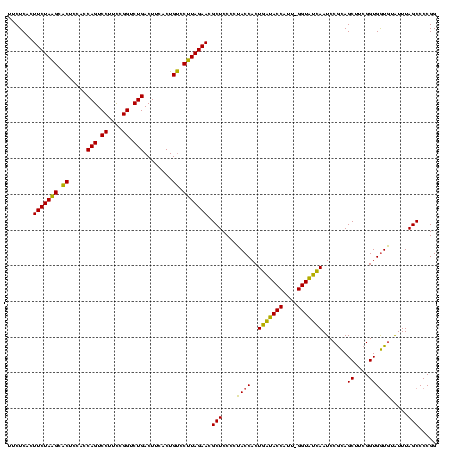
| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.19 |
| Mean single sequence MFE | -38.83 |
| Consensus MFE | -38.54 |
| Energy contribution | -35.67 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.56 |
| SVM RNA-class probability | 0.964056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/1680-1800 CCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACC-AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAA (((.((((..............)))).)))(((((((((-...))))))))).........((.(((((((.((......(((((.((....)).)))))...)).)))))))))..... ( -35.54) >Bcere.0 4651066 120 + 5224283/1680-1800 CCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACCGAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAA (((.((((..............)))).)))(((((((((....))))))))).........((.(((((((.((......(((((.((....)).)))))...)).)))))))))..... ( -36.74) >Bhalo.0 4174298 119 - 4202352/1680-1800 CCGGGGCUAAACGUAUCACCGAAGCUAUGGAUUCGCACC-GUUGGUGCGAGUGGUAGGGGAGCGUUCUACGUGCAGCGAAGUCAGACCGAGAGGACUGGUGGAGCGCGUAGAAGUGAGAA .....(((...(.((((.(((......)))(((((((((-...))))))))))))).)..))).((((((((((..((...((((.((....)).))))))..))))))))))....... ( -44.20) >consensus CCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACC_AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAA .....(((...(..(((.(((......)))(((((((((....))))))))))))..)..))).(((((((.((......(((((.((....)).)))))...)).)))))))....... (-38.54 = -35.67 + -2.88)

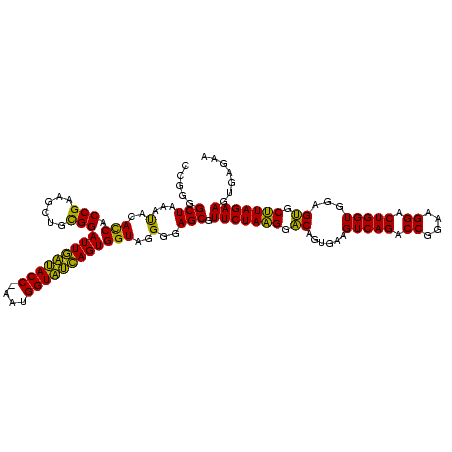

| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -33.13 |
| Consensus MFE | -31.42 |
| Energy contribution | -29.77 |
| Covariance contribution | -1.66 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.95 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523662 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/1720-1840 UUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU-GGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACUAGUGAGCUAUUA (((((((((...((...((((...(((((((((((((...-)))))))...((......))))))))......((.((((.......)))).)).))))..)).))).))))))...... ( -32.40) >Bcere.0 4651066 120 + 5224283/1720-1840 UUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUCGGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACUAGUGAGCUAUUA (((((((((...((...((((...(((((((((((((....)))))))...((......))))))))......((.((((.......)))).)).))))..)).))).))))))...... ( -34.00) >Bhalo.0 4174298 119 - 4202352/1720-1840 UUCGCUGCACGUAGAACGCUCCCCUACCACUCGCACCAAC-GGUGCGAAUCCAUAGCUUCGGUGAUACGUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUA (((((((...((((.........))))...(((((((...-)))))))..........((((((((.(.....((.((((.......)))).)).).))))).))).)))))))...... ( -33.00) >consensus UUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU_GGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACUAGUGAGCUAUUA (((((((......((..((((...(((((((((((((....)))))))...(........)))))))......((.((((.......)))).)).))))...))...)))))))...... (-31.42 = -29.77 + -1.66)

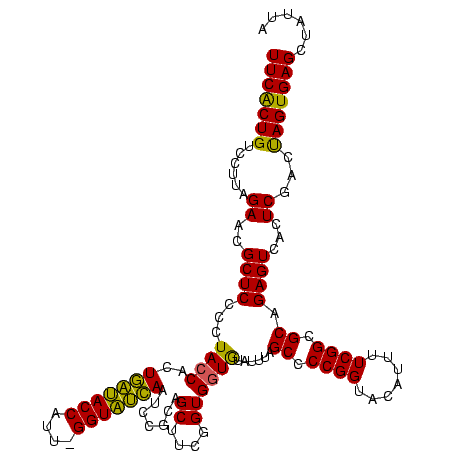
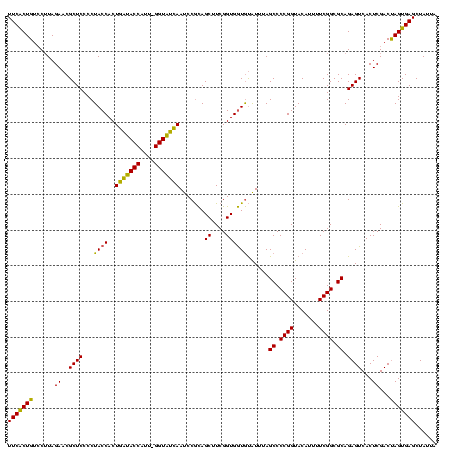
| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -39.16 |
| Consensus MFE | -37.31 |
| Energy contribution | -36.21 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726679 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/1720-1840 UAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACC-AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAA .......(((((.((((((((.((((.((((.........(((.((((..............)))).)))(((((((((-...)))))))))))).).)))))))))....)))))))). ( -38.14) >Bcere.0 4651066 120 + 5224283/1720-1840 UAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACCGAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAA .......(((((.((((((((.((((.((((.........(((.((((..............)))).)))(((((((((....)))))))))))).).)))))))))....)))))))). ( -39.34) >Bhalo.0 4174298 119 - 4202352/1720-1840 UAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAUCACCGAAGCUAUGGAUUCGCACC-GUUGGUGCGAGUGGUAGGGGAGCGUUCUACGUGCAGCGAA .....(((((((((....((..((((((............(((.((((...((......)).)))).)))(((((((((-...))))))))).))))))..))...))).))).)))... ( -40.00) >consensus UAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACC_AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAA .......(((((.((((((((.((((.((((.........(((.((((..............)))).)))(((((((((....)))))))))))).).)))))))))....)))))))). (-37.31 = -36.21 + -1.10)

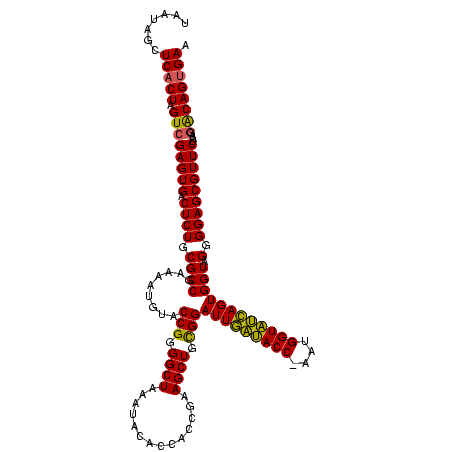
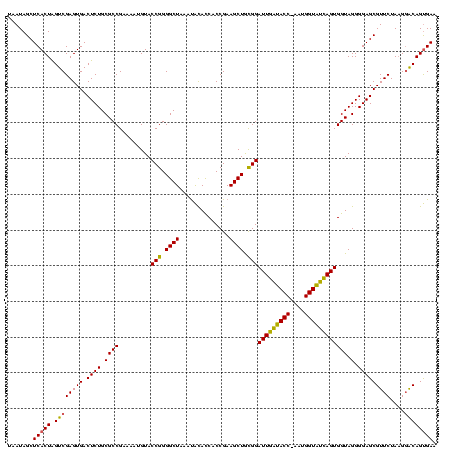
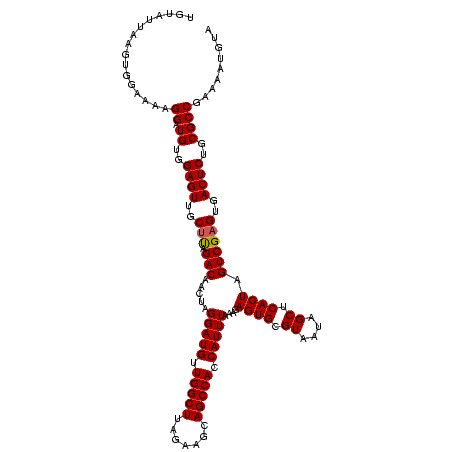
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -36.57 |
| Consensus MFE | -36.12 |
| Energy contribution | -36.23 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.954037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/1800-1920 UGUAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUA ................((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..))))......... ( -37.10) >Bcere.0 4651066 120 + 5224283/1800-1920 UGUAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUA ................((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..))))......... ( -37.10) >Bhalo.0 4174298 120 - 4202352/1800-1920 UACGUUAAGUGGAAAAGGAUGUGGAGUUGCCCAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUA ((((((...(....).((.((..((((..(...((...((((((((.(((((.......))))).))))....((((........)))).))))))..)..))))..))))...)))))) ( -35.50) >consensus UGUAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUA ................((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..))))......... (-36.12 = -36.23 + 0.11)

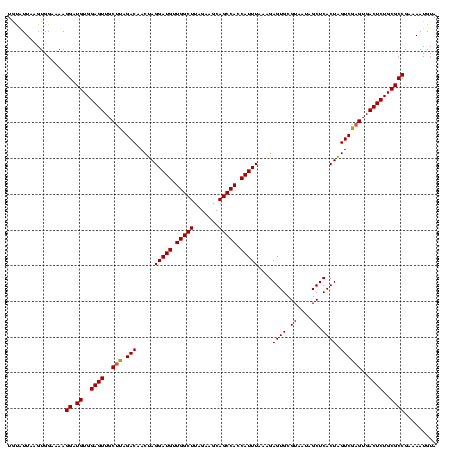
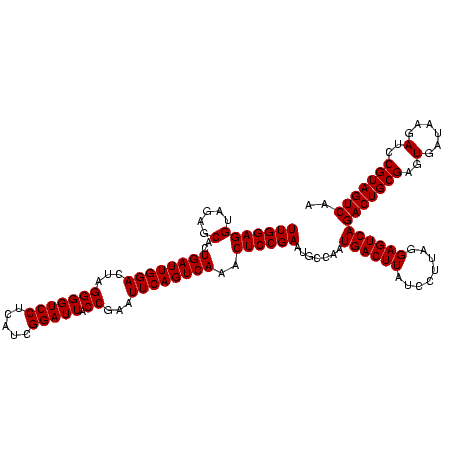
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -36.83 |
| Energy contribution | -36.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/1960-2080 UUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCUCAUCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCAGACUGCGAGUGAUAAGAUCCGUAGUCAA ((((((.....(.(((((..((((.....))))...((((....))))..))))))...)))))).......((((((........))))))(((((((..(......)..))))))).. ( -37.30) >Bcere.0 4651066 120 + 5224283/1960-2080 UUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCUCAUCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCAGACUGCGAGUGAUAAGAUCCGUAGUCAA ((((((.....(.(((((..((((.....))))...((((....))))..))))))...)))))).......((((((........))))))(((((((..(......)..))))))).. ( -37.30) >Bhalo.0 4174298 120 - 4202352/1960-2080 UUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCCCACCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUCGGGAGUCAGACUGCGAGUGCUAAGAUCCGUAGUCAA (((((((.....)..((((((((...(((((((.....))))).))...))))))))..)))))).......((((((........))))))(((((((..(......)..))))))).. ( -36.50) >consensus UUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCUCAUCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCAGACUGCGAGUGAUAAGAUCCGUAGUCAA (((((((.....)..((((((((...(((((((.....))))).))...))))))))..)))))).......((((((........))))))(((((((..(......)..))))))).. (-36.83 = -36.83 + 0.00)

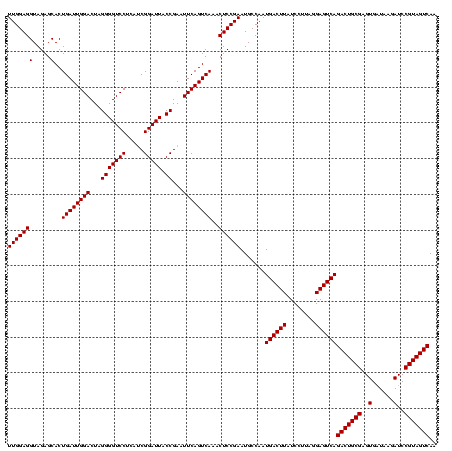
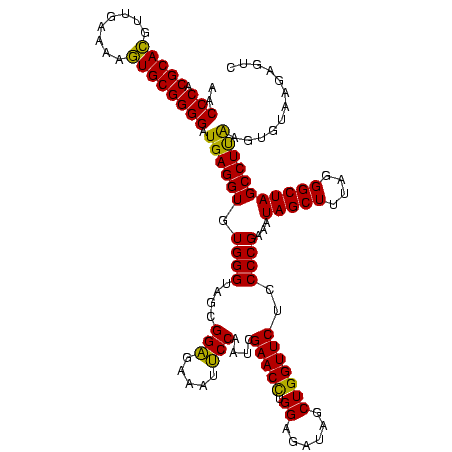
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -38.67 |
| Consensus MFE | -38.88 |
| Energy contribution | -37.33 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.37 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2080-2200 AACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAAGAGUC ..(((.(((((........)))))))).((((((.((((....((((...))))....(((((.((......)))))))..))))...(((((....)))))))))))............ ( -38.40) >Bcere.0 4651066 120 + 5224283/2080-2200 AACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAAGAGUC ..(((.(((((........)))))))).((((((.((((....((((...))))....(((((.((......)))))))..))))...(((((....)))))))))))............ ( -38.40) >Bhalo.0 4174298 119 - 4202352/2080-2200 AACCCACGCAUGUUGAAAAAUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACUCGGAAAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUCGAG-GGAAGAGUC ..(((.(((((........)))))))).((((((.......((((.(((..((...(((....))).....))...))).))))....(((((....)))))))))))..-......... ( -39.20) >consensus AACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAAGAGUC ..(((.(((((........)))))))).((((((.((((....(((.....)))....(((((.((......)))))))..))))...(((((....)))))))))))............ (-38.88 = -37.33 + -1.55)

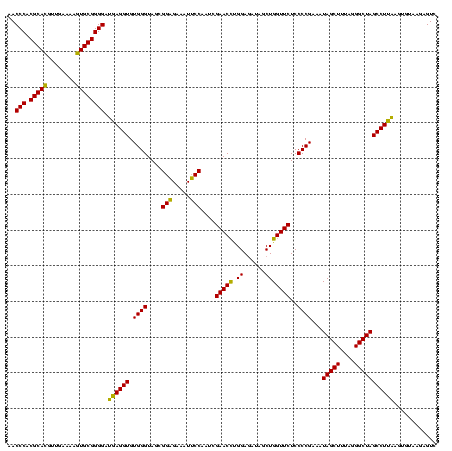
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -31.87 |
| Consensus MFE | -30.03 |
| Energy contribution | -30.03 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.07 |
| SVM RNA-class probability | 0.909533 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2120-2240 AGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACA ....((((........((((((((..(((.....)))................((((((((........))))).))).))))))))...(((((......))))).......))))... ( -33.90) >Bcere.0 4651066 120 + 5224283/2120-2240 AGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACA ....((((........((((((((..(((.....)))................((((((((........))))).))).))))))))...(((((......))))).......))))... ( -33.90) >Bhalo.0 4174298 120 - 4202352/2120-2240 AGAACCAGCUAUUUCCGAGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCAUUUUUCAACAUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACA ....((((........(((((((..(((((((...(((..((((((...........((((........))))))))))..))).........)))))))..)))))))....))))... ( -27.80) >consensus AGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACA ....((((........((((((((..(((.....)))................((((((((........))))).))).))))))))...(((((......))))).......))))... (-30.03 = -30.03 + 0.00)

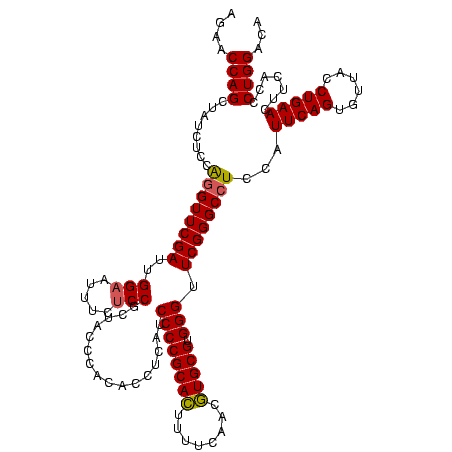
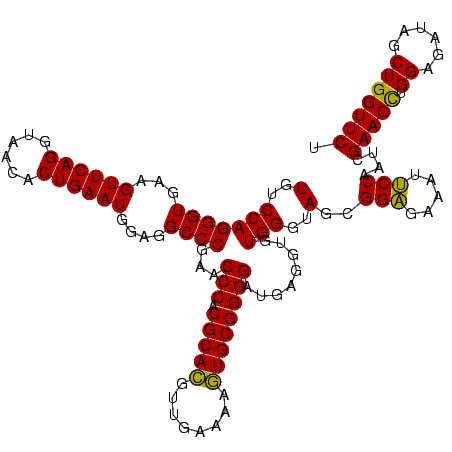
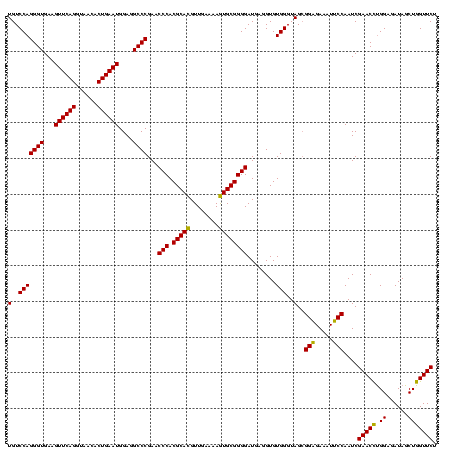
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -40.17 |
| Consensus MFE | -39.64 |
| Energy contribution | -38.53 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824861 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2120-2240 UGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCU ..((((((.(((.((((((......))))))((((.((((.((((((((.(((........)))........))))))))..))).)...))))..)))..))))))............. ( -40.90) >Bcere.0 4651066 120 + 5224283/2120-2240 UGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCU ..((((((.(((.((((((......))))))((((.((((.((((((((.(((........)))........))))))))..))).)...))))..)))..))))))............. ( -40.90) >Bhalo.0 4174298 120 - 4202352/2120-2240 UGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCAUGUUGAAAAAUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACUCGGAAAUAGCUGGUUCU (..(((((((...((((((......))))))....))))...(((.(((((........))))))))........)))..)..(((.....)))....((((.(((......))))))). ( -38.70) >consensus UGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCU (..(((((((...((((((......))))))....))))...(((.(((((........))))))))........)))..)..(((.....)))....(((((.((......))))))). (-39.64 = -38.53 + -1.11)

| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -36.23 |
| Consensus MFE | -35.21 |
| Energy contribution | -34.77 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682275 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2160-2280 ACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCACGUACUA (((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))........((((..((((((...)))).)).)))) ( -37.30) >Bcere.0 4651066 120 + 5224283/2160-2280 ACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCACGUACUA (((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))........((((..((((((...)))).)).)))) ( -37.30) >Bhalo.0 4174298 120 - 4202352/2160-2280 ACCCACACCUCAUCCCCGCAUUUUUCAACAUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACACGGUUUCGGGUCUACGACGGCGUACUA (((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).))))).......(((.(.(((......))).).))).... ( -34.10) >consensus ACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUACGACCACGUACUA ((((..(((.(((.(((((((........))))).))(((((((......(((((......)))))......)))))))))))))(((((....))))).)))).((((....))))... (-35.21 = -34.77 + -0.44)

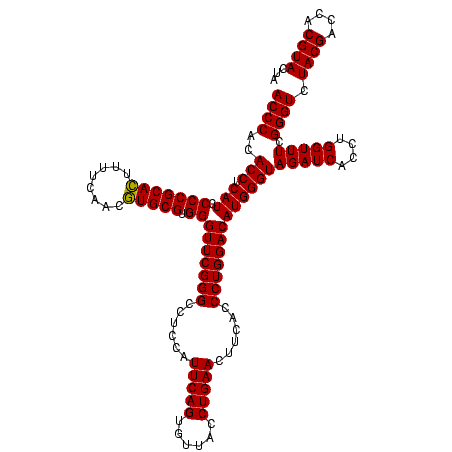
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -46.07 |
| Consensus MFE | -44.95 |
| Energy contribution | -44.40 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2160-2280 UAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGU ...(((.((((((......)))..))).)))....((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))) ( -47.30) >Bcere.0 4651066 120 + 5224283/2160-2280 UAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGU ...(((.((((((......)))..))).)))....((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))) ( -47.30) >Bhalo.0 4174298 120 - 4202352/2160-2280 UAGUACGCCGUCGUAGACCCGAAACCGUGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCAUGUUGAAAAAUGCGGGGAUGAGGUGUGGGU ...(((((..(((......)))....)))))....((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))) ( -43.60) >consensus UAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGU ...(((.((((((......)))..))).)))....((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))) (-44.95 = -44.40 + -0.55)


| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -40.49 |
| Energy contribution | -40.27 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.37 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975867 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2360-2480 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAU ....((((((...((((..(.(....).).((((....))))...........)))).....)).))))(((((((..((((..(((((....)))))..))))......)))))))... ( -41.90) >Bcere.0 4651066 120 + 5224283/2360-2480 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAU ....((((((...((((..(.(....).).((((....))))...........)))).....)).))))(((((((..((((..(((((....)))))..))))......)))))))... ( -41.90) >Bhalo.0 4174298 120 - 4202352/2360-2480 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAAAGAUCCUGAAACCGUGUGCCUACAACUAGUUGGAGCCCAUUAACGGGUGACAGCGUGCCUUUUGUAGAAU .......((....))(((((((.....(((((((....))))((.((......)))).........))).........((((..((((......))))..))))..)))))))....... ( -38.00) >consensus ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAU ....((((((...((((..(.(....).).((((....))))...........)))).....)).))))(((((((..((((..(((((....)))))..))))......)))))))... (-40.49 = -40.27 + -0.22)

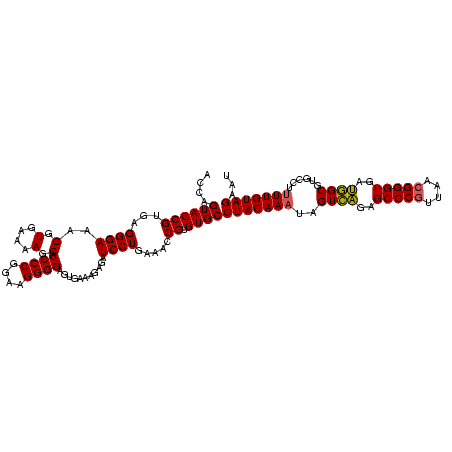
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -33.43 |
| Energy contribution | -31.00 |
| Covariance contribution | -2.43 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.929318 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2560-2680 ACUCAGGAUACAUUCAAGAGAGAACGAAGUUUCGACUACGGGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUA ....((((..(((((.((((.((((((.((...(((...(((((....)))))....((.((((((....))))).).))))).)).))))))))))(((......))).))))))))). ( -34.30) >Bcere.0 4651066 120 + 5224283/2560-2680 ACUCAGGAUACAUUCAAGAGAGAACGAAGUUUCGACUACGGGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUA ....((((..(((((.((((.((((((.((...(((...(((((....)))))....((.((((((....))))).).))))).)).))))))))))(((......))).))))))))). ( -34.30) >Bhalo.0 4174298 120 - 4202352/2560-2680 ACUCAGGAUCCACUCUGGAGGAAACGAAGUUUCGGCUACAGGGUUGUUACCUUCUGUGACGGACCUUUCCAGGUCGCUUCACCUACUUCGUUUCUUUCUGACUCCGUAUAGAGUGUCCUA ....((((..((((((((((((((((((((...(((((((((((....)))..)))))...(((((....))))))))......)))))))))))))).(....)....)))))))))). ( -42.10) >consensus ACUCAGGAUACAUUCAAGAGAGAACGAAGUUUCGACUACGGGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUA ....((((..(((((.((((.((((((.((.........(((((....)))))..((((.((((((....))))).).))))..)).))))))))))...((...))...))))))))). (-33.43 = -31.00 + -2.43)


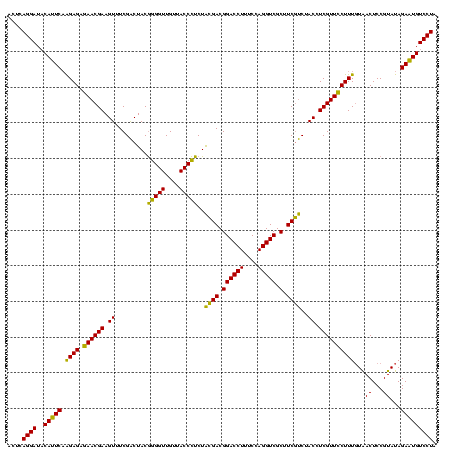
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.00 |
| Mean single sequence MFE | -43.74 |
| Consensus MFE | -43.22 |
| Energy contribution | -40.34 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.996721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2560-2680 UAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCCCGUAGUCGAAACUUCGUUCUCUCUUGAAUGUAUCCUGAGU ((((((((((.....((((.......((((((((((.(((((.(.(((((....)))))).))...(.((((....)))))...)))...)))))))))))))).)))))..)))))... ( -41.91) >Bcere.0 4651066 120 + 5224283/2560-2680 UAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCCCGUAGUCGAAACUUCGUUCUCUCUUGAAUGUAUCCUGAGU ((((((((((.....((((.......((((((((((.(((((.(.(((((....)))))).))...(.((((....)))))...)))...)))))))))))))).)))))..)))))... ( -41.91) >Bhalo.0 4174298 120 - 4202352/2560-2680 UAGGACACUCUAUACGGAGUCAGAAAGAAACGAAGUAGGUGAAGCGACCUGGAAAGGUCCGUCACAGAAGGUAACAACCCUGUAGCCGAAACUUCGUUUCCUCCAGAGUGGAUCCUGAGU (((((((((((....((((.......((((((((((..((((.(.(((((....)))))).))))....(((.(((....))).)))...))))))))))))))))))))..)))))... ( -47.41) >consensus UAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCCCGUAGUCGAAACUUCGUUCUCUCUUGAAUGUAUCCUGAGU ((((((((((.....((((.......((((((((((..((((.(.(((((....)))))).))))...(((.......))).........)))))))))))))).)))))..)))))... (-43.22 = -40.34 + -2.88)


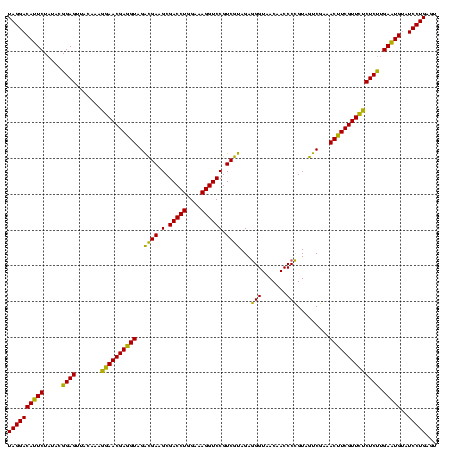
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -33.74 |
| Energy contribution | -32.63 |
| Covariance contribution | -1.11 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2600-2720 GGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUACAACCCCAAGAGGCAAGCCUCUUGGUUUGGGCUAUGUUCC ((((....))))...((((.((((((....))))).).))))............((((((......))))))..((((.......((((((((....))))))))...))))........ ( -33.50) >Bcere.0 4651066 120 + 5224283/2600-2720 GGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUACAACCCCAAGAGGCAAGCCUCUUGGUUUGGGCUAUGUUCC ((((....))))...((((.((((((....))))).).))))............((((((......))))))..((((.......((((((((....))))))))...))))........ ( -33.50) >Bhalo.0 4174298 119 - 4202352/2600-2720 GGGUUGUUACCUUCUGUGACGGACCUUUCCAGGUCGCUUCACCUACUUCGUUUCUUUCUGACUCCGUAUAGAGUGUCCUACAACCCCAAGAGGCAAGCCUCUUGGUUUGGGCUGU-UUCC (((((((.((.(((((((.((((.......((((......)))).....(((.......)))))))))))))).))...)))))))(((((((....)))))))...........-.... ( -37.30) >consensus GGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUACAACCCCAAGAGGCAAGCCUCUUGGUUUGGGCUAUGUUCC (((((((.((..(((((((.((((((....))))).).)))).(((...(((.......)))...))).)))..))...)))))))(((((((....)))))))................ (-33.74 = -32.63 + -1.11)

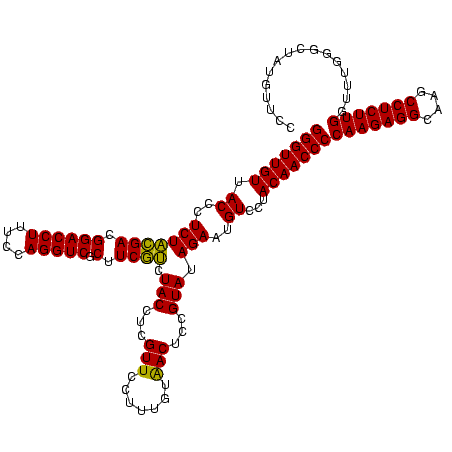
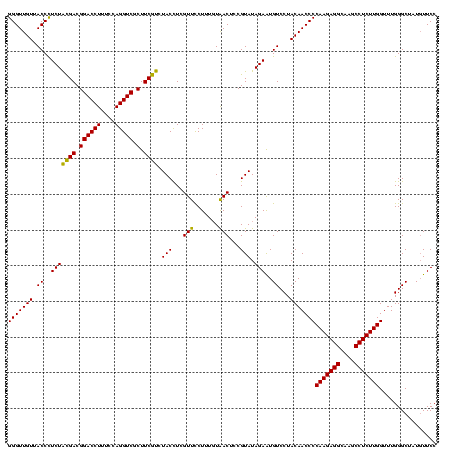
| Location | 4,652,088 – 4,652,208 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -41.57 |
| Consensus MFE | -40.69 |
| Energy contribution | -40.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.83 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 120 + 5227293/2600-2720 GGAACAUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCC ((........))...((((((((....))))))))((((((...((.((((((.....(((.......)))......((((....(((((....))))))))))))))).)).)))))). ( -41.30) >Bcere.0 4651066 120 + 5224283/2600-2720 GGAACAUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCC ((........))...((((((((....))))))))((((((...((.((((((.....(((.......)))......((((....(((((....))))))))))))))).)).)))))). ( -41.30) >Bhalo.0 4174298 119 - 4202352/2600-2720 GGAA-ACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUAUACGGAGUCAGAAAGAAACGAAGUAGGUGAAGCGACCUGGAAAGGUCCGUCACAGAAGGUAACAACCC (...-.)........((((((((....))))))))((((((...((..(((...((...((.....))..))......((((.(.(((((....)))))).)))))))..)).)))))). ( -42.10) >consensus GGAACAUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCC ((........))...((((((((....))))))))((((((...((.((((.(((...(((.......)))...))).((((.(.(((((....)))))).)))))))).)).)))))). (-40.69 = -40.47 + -0.22)

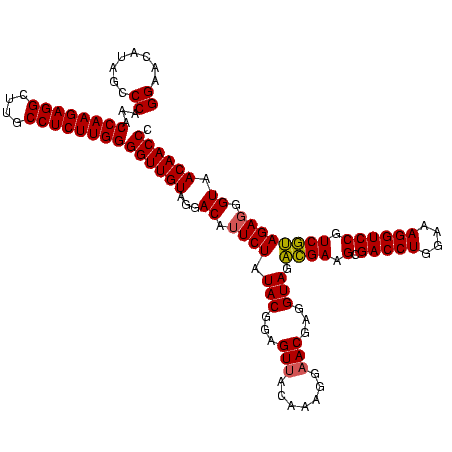

| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -31.65 |
| Energy contribution | -30.10 |
| Covariance contribution | -1.55 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.962858 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/2760-2880 CUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAU ......(((((.(((..((....)))))..)))))(((((...((((-((..((........)).))))))((((((((......))))))))(((((((......)))))))..))))) ( -32.50) >Bcere.0 4651066 119 + 5224283/2760-2880 CUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAU ......(((((.(((..((....)))))..)))))(((((...((((-((..((........)).))))))((((((((......))))))))(((((((......)))))))..))))) ( -32.50) >Bhalo.0 4174298 120 - 4202352/2760-2880 CUUCCUCUGGGUACUUAGAUGUUUCAGUUCCCCAGGUCUGCCUCCUCAUACCCUAUAGAUUCAGGUAUGGGUACCAUCCCAUUACGGAUGGUGGGUUCCCCCAUUCGGAUAUCCCCGGAU ..(((.(((((.(((..((....)))))..)))))(((((....((((((((...........)))))))).(((((((......)))))))(((....)))...)))))......))). ( -37.80) >consensus CUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUCCU_UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAU ..(((((((((.(((..((....)))))..))))))((((.........((((.....((((......))))(((((((......))))))))))).........)))).......))). (-31.65 = -30.10 + -1.55)

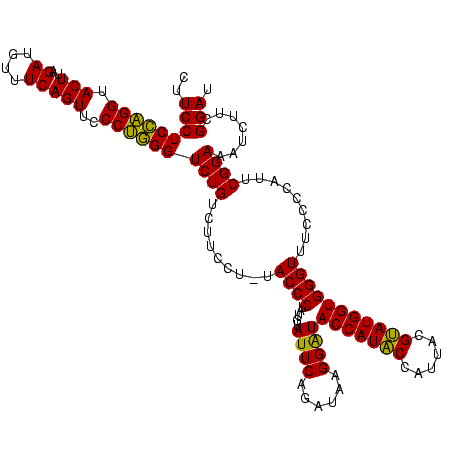
| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -41.10 |
| Consensus MFE | -36.56 |
| Energy contribution | -35.23 |
| Covariance contribution | -1.33 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/2760-2880 AUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAG ..........((((....(((....)))(((((((......))))))).(((((((((.........)))))-))))........(((((..(((..........))).))))).)))). ( -40.20) >Bcere.0 4651066 119 + 5224283/2760-2880 AUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAG ..........((((....(((....)))(((((((......))))))).(((((((((.........)))))-))))........(((((..(((..........))).))))).)))). ( -40.20) >Bhalo.0 4174298 120 - 4202352/2760-2880 AUCCGGGGAUAUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACCCAUACCUGAAUCUAUAGGGUAUGAGGAGGCAGACCUGGGGAACUGAAACAUCUAAGUACCCAGAGGAAG .((((((.....((....(((....)))(((((((......))))))).(((((((((.........))))))).)).))....))).(((.(((..........))).)))...))).. ( -42.90) >consensus AUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA_AGGAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAG .(((..............(((....)))(((((((......))))))).(((((((((.........))))).))))........(((((..(((..........))).))))).))).. (-36.56 = -35.23 + -1.33)

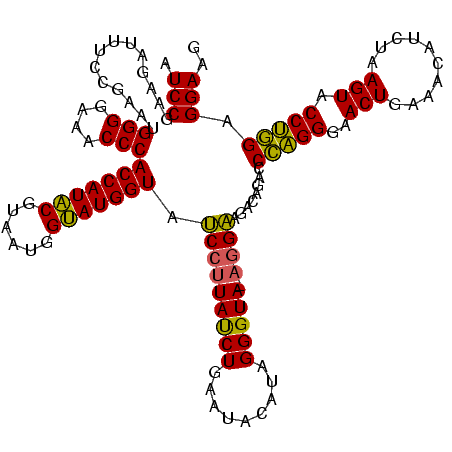
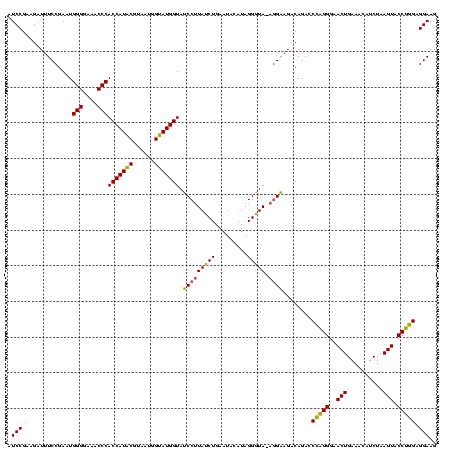
| Location | 4,652,088 – 4,652,207 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.30 |
| Mean single sequence MFE | -43.87 |
| Consensus MFE | -39.60 |
| Energy contribution | -38.50 |
| Covariance contribution | -1.10 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.53 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.06 |
| SVM RNA-class probability | 0.986875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4652088 119 + 5227293/2800-2920 CUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGA .......((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........)))))-))))... ( -42.20) >Bcere.0 4651066 119 + 5224283/2800-2920 CUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGA .......((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........)))))-))))... ( -42.20) >Bhalo.0 4174298 120 - 4202352/2800-2920 CGAACGGCGAAACGCCUCGGGGAGCUGUAAGUAAGCUUUGAUCCGGGGAUAUCCGAAUGGGGGAACCCACCAUCCGUAAUGGGAUGGUACCCAUACCUGAAUCUAUAGGGUAUGAGGAGG ....(((.......((((((((((((.......)))))...)))))))....)))...(((....)))(((((((......))))))).(((((((((.........))))))).))... ( -47.20) >consensus CUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA_AGGAAGA .......((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........))))).))))... (-39.60 = -38.50 + -1.10)

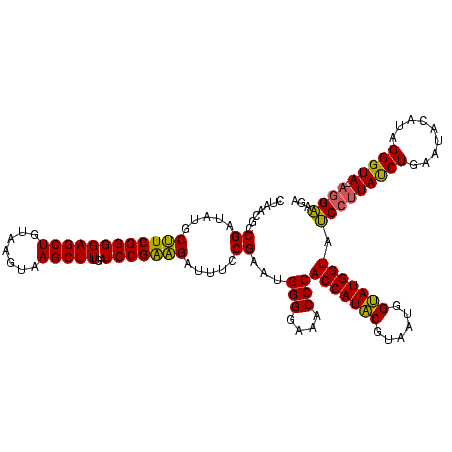
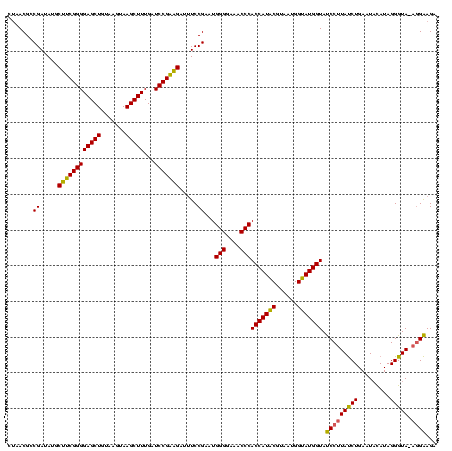
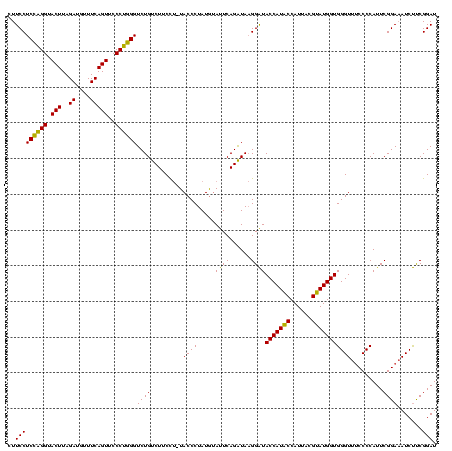
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:15:56 2006