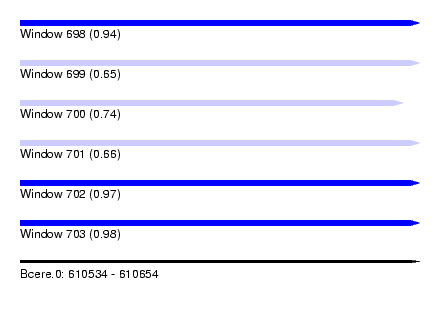
| Sequence ID | Bcere.0 |
|---|---|
| Location | 610,534 – 610,654 |
| Length | 120 |
| Max. P | 0.980991 |

| Location | 610,534 – 610,654 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -39.44 |
| Consensus MFE | -36.84 |
| Energy contribution | -36.27 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.938808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 610534 120 + 5224283/0-120 GGGUUCGGCAGUUCGAAUCUGCCCCCCUCCACCAUUUACAUAGGGGCAUAGUUUAAAGGUAGAACUGAGGUCUCCAAAACCUCCAGUGUGGGUUCGAUUCCUACUGCCCCUGCCAAAUAU .(((..((((((..(((((.((((((((.............))))(((((((((.......)))))(((((.......)))))..))))))))..)))))..))))))...)))...... ( -39.12) >Banth.0 538032 120 + 5227293/0-120 GGGUUCGGCAGUUCGAAUCUGCCCCCCUCCACCAUUUACAUAGGGGCAUAGUUUAAAGGUAGAACUGAGGUCUCCAAAACCUCCAGUGUGGGUUCGAUUCCUACUGCCCCUGCCAAAUAU .(((..((((((..(((((.((((((((.............))))(((((((((.......)))))(((((.......)))))..))))))))..)))))..))))))...)))...... ( -39.12) >Blich.0 926299 120 + 4222334/0-120 GGGUUCGGCGGUUCGAAUCCGUCCCCCUCCACCAUGUUUUUAGGGGCAUAGUUUAACGGUAGAACAGAGGUCUCCAAAACCUCCGGUGUGGGUUCGAUUCCUACUGCCCCUGCCAAUUUU .(((..((((((..(((((.(..(((...((((((((((....)))))).((((.......)))).(((((.......))))).)))).)))..))))))..))))))...)))...... ( -39.20) >Bsubt.0 951691 118 + 4214630/0-120 GGGUUCGGCAGUUCGAAUCUGCCCCCCUCCACCAU--UUAUAGGGGCAUAGUGUAACGGUAGAACAGAGGUCUCCAAAACCUCCGGUGUGGGUUCGAUUCCUACUGCCCCUGCCAAAUUU .(((..((((((..(((((.((((.....((((.(--((((.(..((.....))..).)))))...(((((.......))))).)))).))))..)))))..))))))...)))...... ( -40.30) >consensus GGGUUCGGCAGUUCGAAUCUGCCCCCCUCCACCAUUUACAUAGGGGCAUAGUUUAAAGGUAGAACAGAGGUCUCCAAAACCUCCAGUGUGGGUUCGAUUCCUACUGCCCCUGCCAAAUAU (((...(((((.......))))).))).............((((((((..................(((((.......)))))....(((((.......)))))))))))))........ (-36.84 = -36.27 + -0.56)

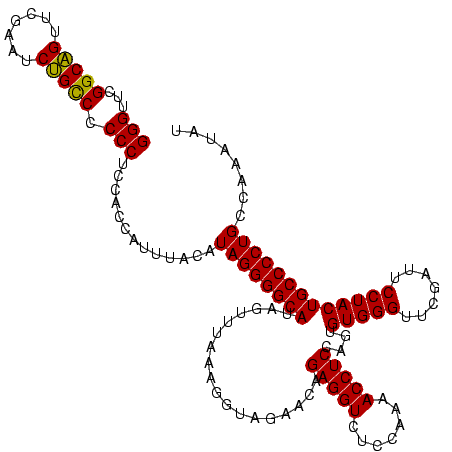
| Location | 610,534 – 610,654 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -40.25 |
| Consensus MFE | -36.91 |
| Energy contribution | -37.41 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.651846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 610534 120 + 5224283/0-120 AUAUUUGGCAGGGGCAGUAGGAAUCGAACCCACACUGGAGGUUUUGGAGACCUCAGUUCUACCUUUAAACUAUGCCCCUAUGUAAAUGGUGGAGGGGGGCAGAUUCGAACUGCCGAACCC ............((((((..((((((..(((..((((.(((((.....)))))))))((((((((((...((......))..)))).)))))).)))..).)))))..))))))...... ( -39.30) >Banth.0 538032 120 + 5227293/0-120 AUAUUUGGCAGGGGCAGUAGGAAUCGAACCCACACUGGAGGUUUUGGAGACCUCAGUUCUACCUUUAAACUAUGCCCCUAUGUAAAUGGUGGAGGGGGGCAGAUUCGAACUGCCGAACCC ............((((((..((((((..(((..((((.(((((.....)))))))))((((((((((...((......))..)))).)))))).)))..).)))))..))))))...... ( -39.30) >Blich.0 926299 120 + 4222334/0-120 AAAAUUGGCAGGGGCAGUAGGAAUCGAACCCACACCGGAGGUUUUGGAGACCUCUGUUCUACCGUUAAACUAUGCCCCUAAAAACAUGGUGGAGGGGGACGGAUUCGAACCGCCGAACCC ....((((((((((((((.((........))))..((((((((.....))))))))................)))))))........(((.((((....)...)))..)))))))).... ( -38.40) >Bsubt.0 951691 118 + 4214630/0-120 AAAUUUGGCAGGGGCAGUAGGAAUCGAACCCACACCGGAGGUUUUGGAGACCUCUGUUCUACCGUUACACUAUGCCCCUAUAA--AUGGUGGAGGGGGGCAGAUUCGAACUGCCGAACCC ............((((((..((((((..(((....((((((((.....))))))))((((((((((................)--))))))))))))..).)))))..))))))...... ( -43.99) >consensus AAAUUUGGCAGGGGCAGUAGGAAUCGAACCCACACCGGAGGUUUUGGAGACCUCAGUUCUACCGUUAAACUAUGCCCCUAUAAAAAUGGUGGAGGGGGGCAGAUUCGAACUGCCGAACCC ............((((((..((((((..(((....((((((((.....))))))))(((((((........................))))))))))..).)))))..))))))...... (-36.91 = -37.41 + 0.50)

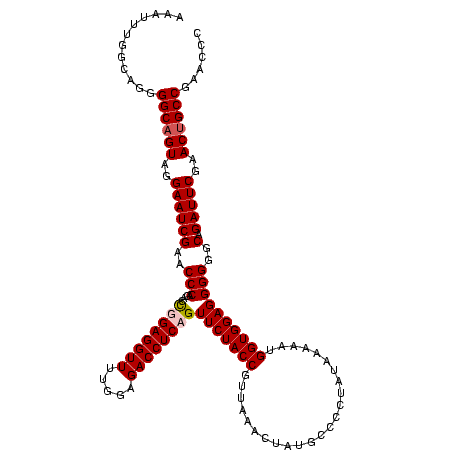
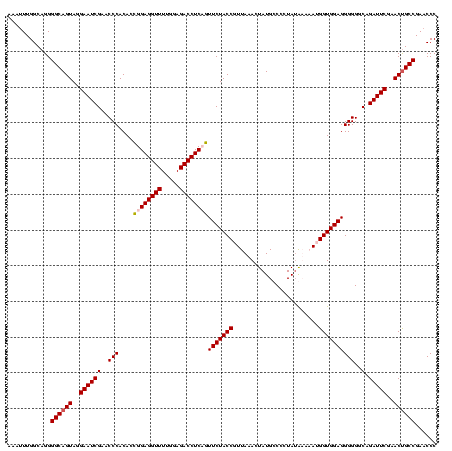
| Location | 610,534 – 610,649 |
|---|---|
| Length | 115 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.66 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -28.50 |
| Energy contribution | -28.75 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 610534 115 + 5224283/40-160 UAACCACUUCGCCACAACCGCCAUGUUGUGUAAA-----UAUAUUUGGCAGGGGCAGUAGGAAUCGAACCCACACUGGAGGUUUUGGAGACCUCAGUUCUACCUUUAAACUAUGCCCCUA ...................((((...((((....-----))))..))))(((((((((.((........))))((((.(((((.....)))))))))...............))))))). ( -31.00) >Banth.0 538032 115 + 5227293/40-160 UAACCACUUCGCCACAACCGCCAUGUUGUGUAAA-----UAUAUUUGGCAGGGGCAGUAGGAAUCGAACCCACACUGGAGGUUUUGGAGACCUCAGUUCUACCUUUAAACUAUGCCCCUA ...................((((...((((....-----))))..))))(((((((((.((........))))((((.(((((.....)))))))))...............))))))). ( -31.00) >Blich.0 926299 116 + 4222334/40-160 UAACCACUUCGCCACAACCGCCAUAAAUUGUGAAU----UAAAAUUGGCAGGGGCAGUAGGAAUCGAACCCACACCGGAGGUUUUGGAGACCUCUGUUCUACCGUUAAACUAUGCCCCUA ...................((((....(((.....----)))...))))(((((((((.((........))))..((((((((.....))))))))................))))))). ( -31.10) >Bsubt.0 951691 120 + 4214630/40-160 UAACCACUUCGCCACAACCGCCAUAAUCUCAAGAUGUCAUAAAUUUGGCAGGGGCAGUAGGAAUCGAACCCACACCGGAGGUUUUGGAGACCUCUGUUCUACCGUUACACUAUGCCCCUA ..........((((...........(((....)))..........))))(((((((((.((........))))..((((((((.....))))))))................))))))). ( -32.60) >consensus UAACCACUUCGCCACAACCGCCAUAAUGUGUAAA_____UAAAUUUGGCAGGGGCAGUAGGAAUCGAACCCACACCGGAGGUUUUGGAGACCUCAGUUCUACCGUUAAACUAUGCCCCUA ...................((((......................))))(((((((((.((........))))..((((((((.....))))))))................))))))). (-28.50 = -28.75 + 0.25)

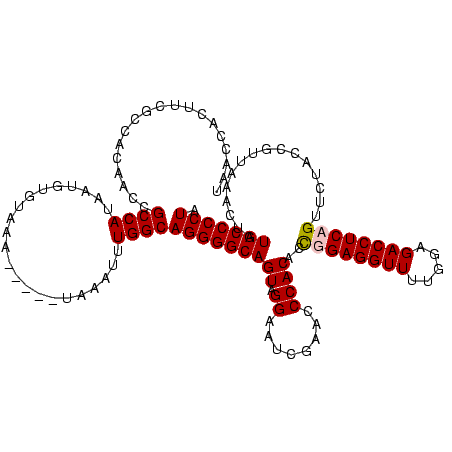
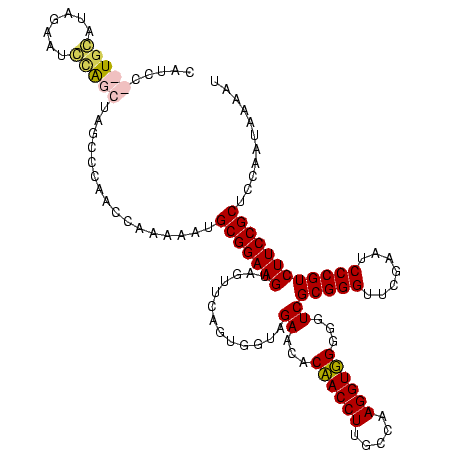
| Location | 610,534 – 610,654 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.27 |
| Mean single sequence MFE | -37.90 |
| Consensus MFE | -30.79 |
| Energy contribution | -31.23 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.657746 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 610534 120 + 5224283/320-440 CAUGCGCUGGUUCGAAUCCAGCUAGCCCAGCCAUUUUUGCGGAAGUAGUUCAGUGGUAGAAUACAACCUUGCCAAGGUUGGGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAGUAAAAU ...(((..(((((((((((.((..((((.((((((.((((....))))...))))))......((((((.....)))))))))).))))))))))))).))).................. ( -43.30) >Banth.0 538032 120 + 5227293/320-440 CAUGCGCUGGUUCGAAUCCAGCUAGCCCAGCCAUUUUUGCGGAAGUAGUUCAGUGGUAGAAUACAACCUUGCCAAGGUUGGGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAGUAAAAU ...(((..(((((((((((.((..((((.((((((.((((....))))...))))))......((((((.....)))))))))).))))))))))))).))).................. ( -43.30) >Blich.0 926299 118 + 4222334/320-440 CAUCC--UGCAUAUGAUAAGAUGAAUAUUAGUUAAAAUGCGGAAGUAGUUCAGUGGUAGAACACCACCUUGCCAAGGUGGGGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAAUUCUUA ((((.--(((....).)).))))...............(((((((..((((.......)))).((((((.....)))))).....(((((.......))))))))))))........... ( -32.40) >Bsubt.0 951691 112 + 4214630/320-440 CAAC---UGCAUAUAAUGUAUUAAGU-CCAUUAAAAAUGCGGAAGUAGUUCAGUGGUAGAACACCACCUUGCCAAGGUGGGGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAAUU---- ....---.((((.(((((........-.)))))...))))(((.((.((((.......)))).((((((.....))))))(((..(((((.......)))))..))))))))....---- ( -32.60) >consensus CAUCC__UGCAUAGAAUCCAGCUAGCCCAACCAAAAAUGCGGAAGUAGUUCAGUGGUAGAACACAACCUUGCCAAGGUGGGGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAAUAAAAU .....(((((.......)))))................(((((((.............((...((((((.....))))))...))(((((.......))))))))))))........... (-30.79 = -31.23 + 0.44)


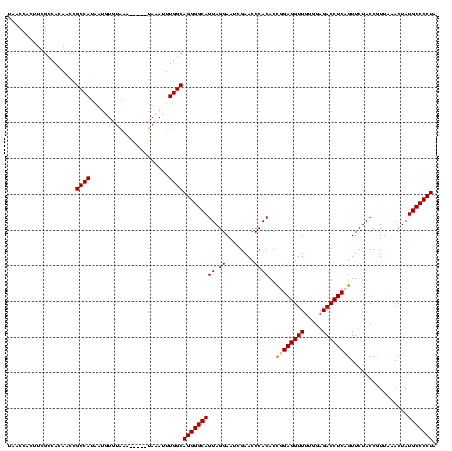
| Location | 610,534 – 610,654 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.71 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -43.12 |
| Energy contribution | -42.38 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.78 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966254 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 610534 120 + 5224283/400-520 GGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAGUAAAAUUUUAUAUGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCACCGGUUCAAAUCCGGUUGCCGCCUUU (((..(((((.......)))))..)))....................(((((((.........(((...((((....))))...)))......(((((.......))))))))))))... ( -42.60) >Banth.0 538032 120 + 5227293/400-520 GGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAGUAAAAUUUUAUAUGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCACCGGUUCAAAUCCGGUUGCCGCCUUU (((..(((((.......)))))..)))....................(((((((.........(((...((((....))))...)))......(((((.......))))))))))))... ( -42.60) >Blich.0 926299 119 + 4222334/400-520 GGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAAUUCUUAAAUG-AUGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCCCCGGUUCGAAUCCGGGUGUCGCCUUA (((((.((((((((((((..((((((((((.....((......)-)((((......))))))))).)))))((((((.......))))))......)))))))))))).)....)))).. ( -44.10) >Bsubt.0 951691 111 + 4214630/400-520 GGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAAUU---------ACGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCCCCGGUUCGAAUCCGGGUGUCGCCUUC (((..(((((.......)))))..))).........---------..(((((((.........(((...((((....))))...)))......(((((.......))))))))))))... ( -42.30) >consensus GGGUCGCGGGUUCGAAUCCCGUCUUCCGCUCCAAUAAAAUUUUA_AUGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCACCGGUUCAAAUCCGGGUGCCGCCUUU (((..(((((.......)))))..)))....................(((((((.........(((...((((....))))...)))......(((((.......))))))))))))... (-43.12 = -42.38 + -0.75)

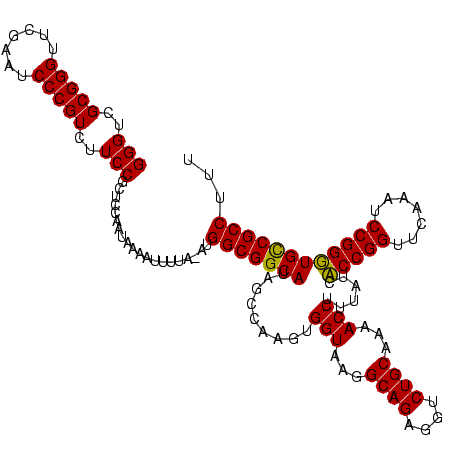
| Location | 610,534 – 610,654 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.32 |
| Mean single sequence MFE | -39.95 |
| Consensus MFE | -40.03 |
| Energy contribution | -39.27 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.92 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.980991 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 610534 120 + 5224283/404-524 CGCGGGUUCGAAUCCCGUCUUCCGCUCCAGUAAAAUUUUAUAUGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCACCGGUUCAAAUCCGGUUGCCGCCUUUUCUU .(((((..((.....))...)))))..................(((((((.........(((...((((....))))...)))......(((((.......))))))))))))....... ( -39.50) >Banth.0 538032 120 + 5227293/404-524 CGCGGGUUCGAAUCCCGUCUUCCGCUCCAGUAAAAUUUUAUAUGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCACCGGUUCAAAUCCGGUUGCCGCCUUUUCUU .(((((..((.....))...)))))..................(((((((.........(((...((((....))))...)))......(((((.......))))))))))))....... ( -39.50) >Blich.0 926299 119 + 4222334/404-524 CGCGGGUUCGAAUCCCGUCUUCCGCUCCAAUUCUUAAAUG-AUGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCCCCGGUUCGAAUCCGGGUGUCGCCUUAUUUC ..((((((((((((..((((((((((.....((......)-)((((......))))))))).)))))((((((.......))))))......))))))))))))((.....))....... ( -41.60) >Bsubt.0 951691 111 + 4214630/404-524 CGCGGGUUCGAAUCCCGUCUUCCGCUCCAAUU---------ACGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCCCCGGUUCGAAUCCGGGUGUCGCCUUCUUAU .(((((..((.....))...))))).......---------..(((((((.........(((...((((....))))...)))......(((((.......))))))))))))....... ( -39.20) >consensus CGCGGGUUCGAAUCCCGUCUUCCGCUCCAAUAAAAUUUUA_AUGGCGGCAUAGCCAAGUGGUAAGGCAGAGGUCUGCAAAACCUUUAUCACCGGUUCAAAUCCGGGUGCCGCCUUUUCUU .(((((..((.....))...)))))..................(((((((.........(((...((((....))))...)))......(((((.......))))))))))))....... (-40.03 = -39.27 + -0.75)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:24:48 2006