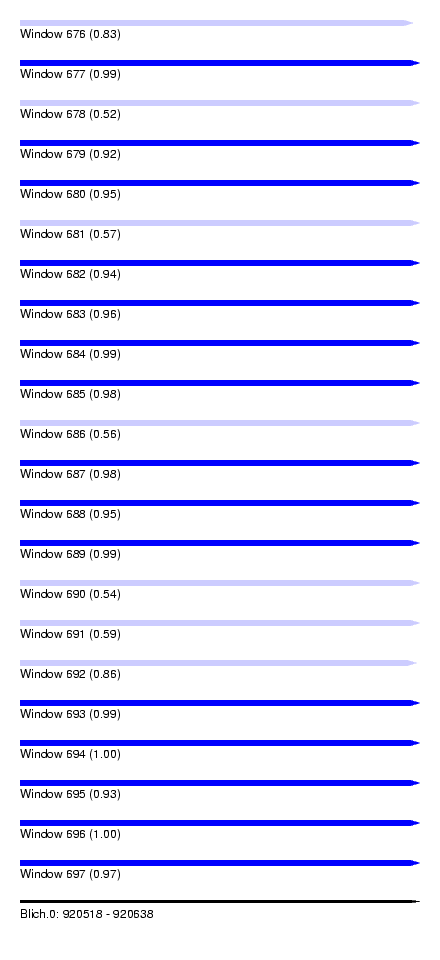
| Sequence ID | Blich.0 |
|---|---|
| Location | 920,518 – 920,638 |
| Length | 120 |
| Max. P | 0.999943 |

| Location | 920,518 – 920,636 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.39 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -41.51 |
| Energy contribution | -40.95 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.830966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 118 + 4222334/40-160 C-GGGAGCUUGCUCCC-UUAGGUCAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGC .-(((((....)))))-...(((((((((((.((((((..((.((......)).))..))))))....((((..((.((((((....)))))).))...)))).))).)).))).))).. ( -46.80) >Bsubt.0 946065 118 + 4214630/40-160 U-GGGAGCUUGCUCCC-UGAUGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGC .-(((((....)))))-........(((((..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).............)))) ( -44.90) >Bcere.0 604964 120 + 5224283/40-160 UUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGC .((((((....))))))........((((.((.(((((................))))))).......((((..((.((((((....)))))).))...)))).............)))) ( -38.39) >Banth.0 532469 120 + 5227293/40-160 UUAAGAGCUUGCUCUUAUGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGC .((((((....))))))...........((((..((((..((.((......)).))..))))......((((..((.((((((....)))))).))...)))).............)))) ( -40.50) >consensus U_AAGAGCUUGCUCCC_UGAAGUUAGCGGCGGACGGGUGAGUAACACGUGGGUAACCUGCCUAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAGUUUGAACCGC .((((((....))))))........(((((..((((((..((.((......)).))..))))))..).((((..((.((((((....)))))).))...)))).............)))) (-41.51 = -40.95 + -0.56)

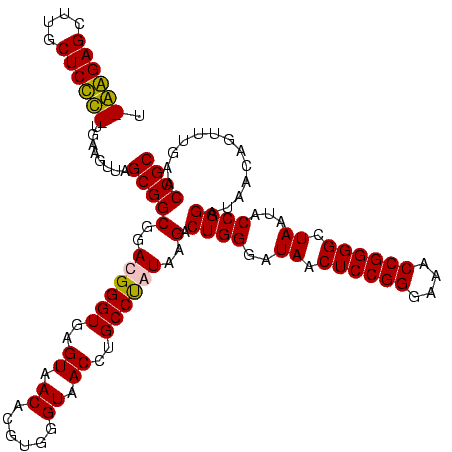
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -46.10 |
| Consensus MFE | -38.40 |
| Energy contribution | -37.40 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.18 |
| Mean z-score | -3.90 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.992804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/80-200 GUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUA ((((((.(..((...))..).)))....((((..((.((((((....)))))).))...)))).))).((((((((((....)))).)))))).....((((((.....))))))..... ( -44.90) >Bsubt.0 946065 118 + 4214630/80-200 GUAACACGUGGGUAACCUGCCUGUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUC-AAACAUAAAAGGUGGCUUC-GCUUACCAAUUA .......((((((((.(..(((......((((..((.((((((....)))))).))...)))).....((((((((((....)))))-)))))....)))..).)).-))))))...... ( -44.00) >Bcere.0 604964 118 + 5224283/80-200 GUAACACGUGGGUAACCUGCCUAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAU-UGAAAGGCGGCUUC-GGCUGUCACUUA .......((((((.....))))))....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))-.....((((((...-.))))))..... ( -46.60) >Banth.0 532469 118 + 5227293/80-200 GUAACACGUGGGUAACCUGCCCAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAU-UGAAAGGCGGCUUC-GGCUGUCACUUA .......((((((.....))))))....((((..((.((((((....)))))).))...)))).....((((((((((....))))))))))-.....((((((...-.))))))..... ( -48.90) >consensus GUAACACGUGGGUAACCUGCCUAUAAGACUGGGAUAACUCCGGGAAACCGGGGCUAAUACCGGAUAACAGUUUGAACCGCAUGGUUCGAAAC_UAAAAGGCGGCUUC_GGCUACCACUUA .......((((((.....))))))....((((..((.((((((....)))))).))...))))......(((((((((....))))).))))......((((((.....))))))..... (-38.40 = -37.40 + -1.00)

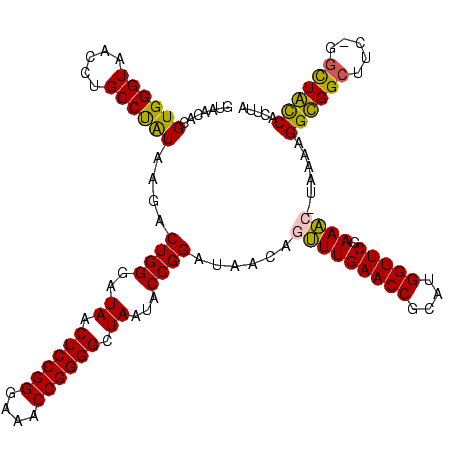
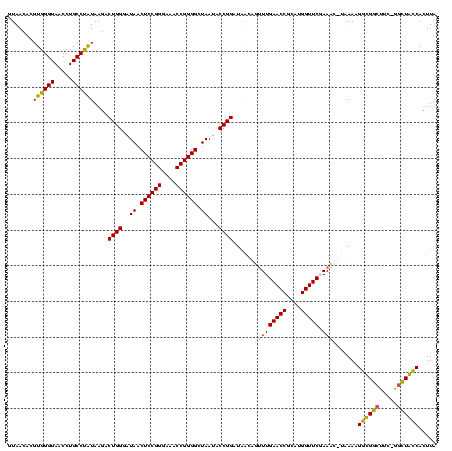
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.69 |
| Mean single sequence MFE | -34.49 |
| Consensus MFE | -26.77 |
| Energy contribution | -27.90 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.78 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.522057 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/80-200 UAAGUGGUAGCUAAAAGCCACCUUUUAUGAUUGGAACCAUGCGGUUCAAUCAAGCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUAC ...(((((((((....(((........((((((.((((....)))))))))).......((...((((.((((....)))).))))..)).........))).))))).))))....... ( -36.00) >Bsubt.0 946065 118 + 4214630/80-200 UAAUUGGUAAGC-GAAGCCACCUUUUAUGUUU-GAACCAUGCGGUUCAAACAACCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUAC .....(((((.(-...(((........(((((-(((((....)))))))))).......((...((((.((((....)))).))))..)).........))).).))))).......... ( -36.90) >Bcere.0 604964 118 + 5224283/80-200 UAAGUGACAGCC-GAAGCCGCCUUUCA-AUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUAGGCAGGUUACCCACGUGUUAC ...(((((((((-...((((.......-((((.(((((....))))).))))......))))..((((.((((....)))).)))).............))).((....))...)))))) ( -32.52) >Banth.0 532469 118 + 5227293/80-200 UAAGUGACAGCC-GAAGCCGCCUUUCA-AUUUCGAACCAUGCGGUUCAAAAUGUUAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUAUGGGCAGGUUACCCACGUGUUAC ...(((..((((-...((((.......-((((.(((((....))))).))))......))))..((((.((((....)))).)))).((((......))))..))))...)))....... ( -32.52) >consensus UAAGUGACAGCC_GAAGCCACCUUUCA_AUUUCGAACCAUGCGGUUCAAAAAAUCAUCCGGUAUUAGCCCCGGUUUCCCGGAGUUAUCCCAGUCUUACAGGCAGGUUACCCACGUGUUAC ...(((((((((....(((..............(((((....)))))............((...((((.((((....)))).))))..)).........))).))))).))))....... (-26.77 = -27.90 + 1.13)

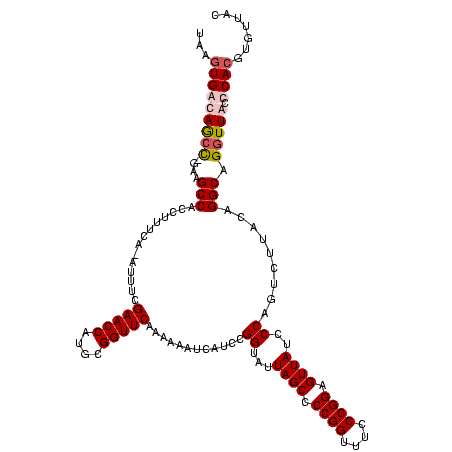
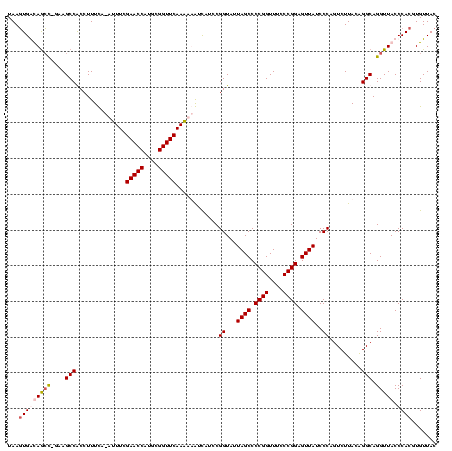
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.57 |
| Mean single sequence MFE | -43.58 |
| Consensus MFE | -32.99 |
| Energy contribution | -31.67 |
| Covariance contribution | -1.31 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.76 |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.923573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/120-240 CGGGAAACCGGGGCUAAUACCGGAUGCUUGAUUGAACCGCAUGGUUCCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAC (((....)))..(((..((((((..(((....((..((((..(((.(((.((.(((..((((((.....))))))..)))..)))))))).))))..))..)))...)))))).)))... ( -41.60) >Bsubt.0 946065 118 + 4214630/120-240 CGGGAAACCGGGGCUAAUACCGGAUGGUUGUUUGAACCGCAUGGUUC-AAACAUAAAAGGUGGCUUC-GCUUACCAAUUACAGAUGGCCCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAC .((....))((((((...(((.......((((((((((....)))))-))))).....)))))))))-.((((((((((((.(((((((....))).)))).).)))))))))))..... ( -48.90) >Bcere.0 604964 118 + 5224283/120-240 CGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAU-UGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAAC (((....)))((((((...(((......((((((((((....))))))))))-.....((((((...-.)))))).....))).))).)))((.((((............)))).))... ( -41.90) >Banth.0 532469 118 + 5227293/120-240 CGGGAAACCGGGGCUAAUACCGGAUAACAUUUUGAACCGCAUGGUUCGAAAU-UGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAAC (((....)))((((((...(((......((((((((((....))))))))))-.....((((((...-.)))))).....))).))).)))((.((((............)))).))... ( -41.90) >consensus CGGGAAACCGGGGCUAAUACCGGAUAACAGUUUGAACCGCAUGGUUCGAAAC_UAAAAGGCGGCUUC_GGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAAC (((....))).(((((((.(((..((...(((((((((....))))).))))......((((((.....)))))).....))..)))....((...)))))))))............... (-32.99 = -31.67 + -1.31)

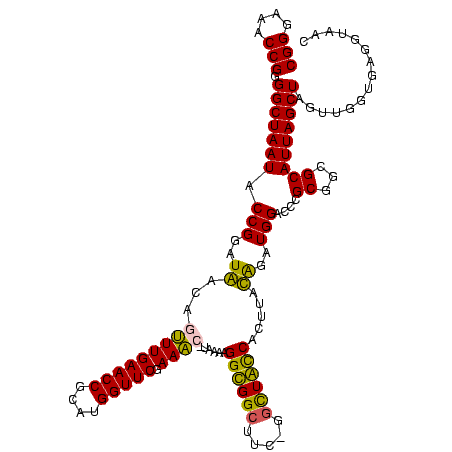
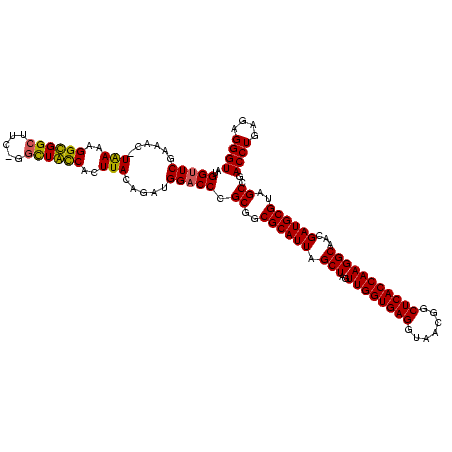
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -45.57 |
| Consensus MFE | -38.39 |
| Energy contribution | -40.20 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.42 |
| SVM RNA-class probability | 0.951834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/160-280 AUGGUUCCAAUCAUAAAAGGUGGCUUUUAGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCGACGAUGCGUAGCCGACCUGAGAGGGU ..((.((((.((.(((..((((((.....))))))..)))..)))))).))((.(((((((.(((..((((((((.......)))))))))))...))))))).))..((((....)))) ( -48.30) >Bsubt.0 946065 118 + 4214630/160-280 AUGGUUC-AAACAUAAAAGGUGGCUUC-GCUUACCAAUUACAGAUGGCCCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGU .......-..........(((((((.(-.((((((((((((.(((((((....))).)))).).))))))))))))....))).))))..(((.(((...))).))).((((....)))) ( -41.80) >Bcere.0 604964 118 + 5224283/160-280 AUGGUUCGAAAU-UGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGU ..((((((....-(((..((((((...-.))))))..)))....))))))(((((((.....(((..((((((((.......)))))))))))..)))))))......((((....)))) ( -46.10) >Banth.0 532469 118 + 5227293/160-280 AUGGUUCGAAAU-UGAAAGGCGGCUUC-GGCUGUCACUUAUGGAUGGACCCGCGUCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGU ..((((((....-(((..((((((...-.))))))..)))....))))))(((((((.....(((..((((((((.......)))))))))))..)))))))......((((....)))) ( -46.10) >consensus AUGGUUCGAAAC_UAAAAGGCGGCUUC_GGCUACCACUUACAGAUGGACCCGCGGCGCAUUAGCUAGUUGGUGAGGUAACGGCUCACCAAGGCAACGAUGCGUAGCCGACCUGAGAGGGU ..(((((......(((..((((((.....))))))..))).....))))).((..((((((.(((..((((((((.......)))))))))))...))))))..))..((((....)))) (-38.39 = -40.20 + 1.81)

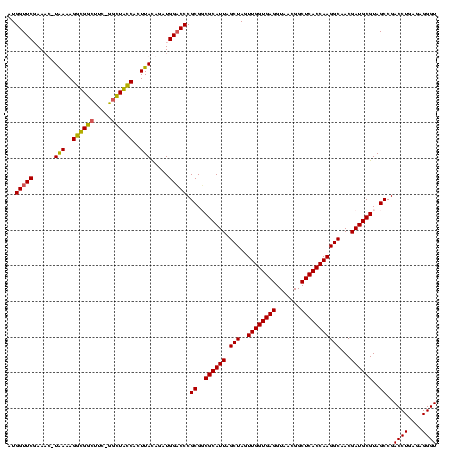
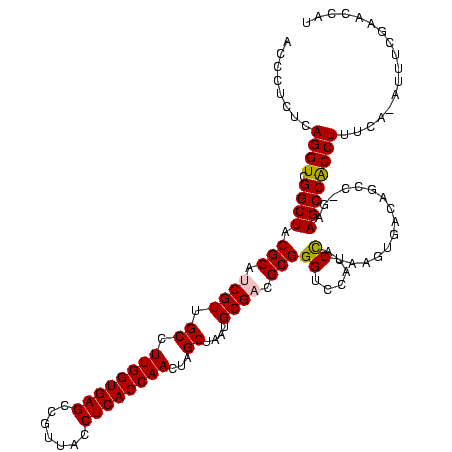
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.50 |
| Mean single sequence MFE | -36.65 |
| Consensus MFE | -29.52 |
| Energy contribution | -29.52 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/160-280 ACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUGUAAGUGGUAGCUAAAAGCCACCUUUUAUGAUUGGAACCAU .........(((.(((.(((...))).)))((((((((.......))))))))...((....)))))..((.((((..((((((.(((.((.....)).))).))))))..)))).)).. ( -37.50) >Bsubt.0 946065 118 + 4214630/160-280 ACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGGCCAUCUGUAAUUGGUAAGC-GAAGCCACCUUUUAUGUUU-GAACCAU ........((((.((((.(((.((((.((.((((((((.......))))))))...((....)))).)))).((((........))))..))-).)))))))).........-....... ( -38.10) >Bcere.0 604964 118 + 5224283/160-280 ACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCC-GAAGCCGCCUUUCA-AUUUCGAACCAU ........(((.(((((.(((.((((.((.((((((((.......))))))))...))....)))).)))(((.(((......)))...)))-..))))))))....-............ ( -35.50) >Banth.0 532469 118 + 5227293/160-280 ACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCC-GAAGCCGCCUUUCA-AUUUCGAACCAU ........(((.(((((.(((.((((.((.((((((((.......))))))))...))....)))).)))(((.(((......)))...)))-..))))))))....-............ ( -35.50) >consensus ACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCAUAAGUGACAGCC_GAAGCCACCUUUCA_AUUUCGAACCAU ........((((.((((.(((.((((.((.((((((((.......))))))))...))....)))).)))((.....))................))))))))................. (-29.52 = -29.52 + 0.00)

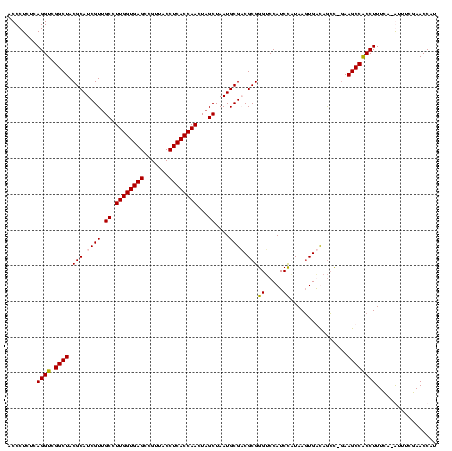
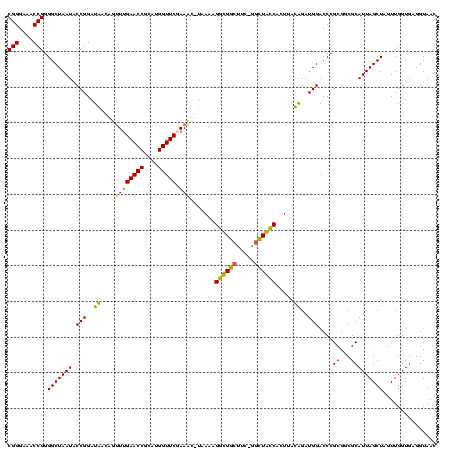
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.50 |
| Mean single sequence MFE | -50.68 |
| Consensus MFE | -47.97 |
| Energy contribution | -48.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.31 |
| SVM RNA-class probability | 0.940133 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/200-320 UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUCGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGUCCAUCUG ..(((...((((((..............((((((((((((.........))))))))))))..(((.((.((((((((.......))))))))...((....)))).)))))))))))). ( -48.00) >Bsubt.0 946065 120 + 4214630/200-320 UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGCCGCGGGGCCAUCUG ...(((.(........).))).(((((.((((((((((((.........))))))))))))...((.((.((((((((.......))))))))...((....)))).)))))))...... ( -47.90) >Bcere.0 604964 120 + 5224283/200-320 UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCA ..(((...(((((((((((.((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...)).))))).))))))))). ( -53.40) >Banth.0 532469 120 + 5227293/200-320 UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCA ..(((...(((((((((((.((......((((((((((((.........))))))))))))...((((..((((((((.......)))))))).))))...)).))))).))))))))). ( -53.40) >consensus UAGGAGUCUGGGCCGUGUCUCAGUCCCAGUGUGGCCGAUCACCCUCUCAGGUCGGCUACGCAUCGUUGCCUUGGUGAGCCGUUACCUCACCAACUAGCUAAUGCGACGCGGGUCCAUCCA ..(((...(((((((((((.((......((((((((((((.........))))))))))))......((.((((((((.......))))))))...))...)).))))).))))))))). (-47.97 = -48.48 + 0.50)

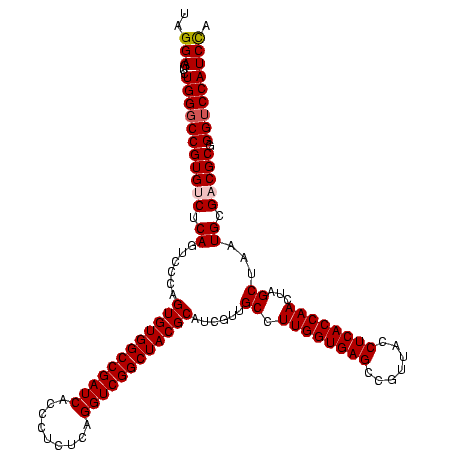
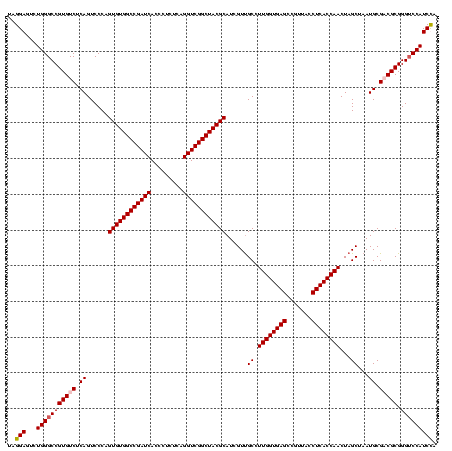
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.61 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -38.81 |
| Energy contribution | -37.12 |
| Covariance contribution | -1.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.55 |
| SVM RNA-class probability | 0.963327 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/360-480 UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCA ((((((((((...((.(((....))).))..((((....))))......)))))).((((((......((((..((((((.....))))))...))))......))))))))))...... ( -35.30) >Bsubt.0 946065 120 + 4214630/360-480 UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGUUUUCGGAUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCA (((((((((((..((.(((....))).))..((((....)))).....))))))).((((((......((((((((((((.....)))))))).))))......))))))))))...... ( -43.90) >Bcere.0 604964 120 + 5224283/360-480 UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCA .............((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))). ( -41.80) >Banth.0 532469 120 + 5227293/360-480 UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGGUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCA .............((.(((....))).))..(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))). ( -41.80) >consensus UCUGACGGAGCAACGCCGCGUGAGUGAUGAAGGCUUUCGGAUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCAGUCGAAUAAGCCGGCACCUUGACGGUACCUAACCAGAAAGCCA .((.(((..((...))..))).)).......(((((((((((......))))....((((((......((((((((((((.....)))))))).))))......))))))..))))))). (-38.81 = -37.12 + -1.69)

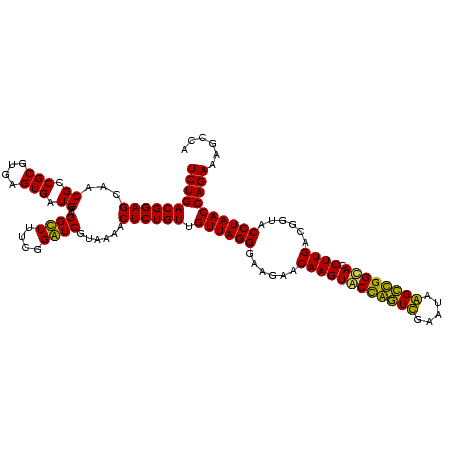
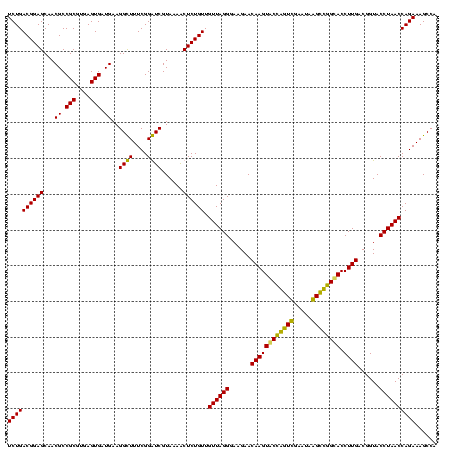
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -41.53 |
| Energy contribution | -39.77 |
| Covariance contribution | -1.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.11 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.985124 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/400-520 AUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG ..........((((..((((((......((((..((((((.....))))))...))))......)))))))))).(((....)))..(((((((((.((....)))))...))))))... ( -36.80) >Bsubt.0 946065 120 + 4214630/400-520 AUCGUAAAGCUCUGUUGUUAGGGAAGAACAAGUACCGUUCGAAUAGGGCGGUACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG ..........((((..((((((......((((((((((((.....)))))))).))))......)))))))))).(((....)))..(((((((((.((....)))))...))))))... ( -41.00) >Bcere.0 604964 120 + 5224283/400-520 GUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG (((((.....((((..((((((......((((((((((((.....)))))))).))))......)))))))))).....)))))...(((((((((.((....)))))...))))))... ( -42.50) >Banth.0 532469 120 + 5227293/400-520 GUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUGCUAGUUGAAUAAGCUGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG (((((.....((((..((((((......((((((((((((.....)))))))).))))......)))))))))).....)))))...(((((((((.((....)))))...))))))... ( -42.50) >consensus AUCGUAAAACUCUGUUGUUAGGGAAGAACAAGUACCAGUCGAAUAAGCCGGCACCUUGACGGUACCUAACCAGAAAGCCACGGCUAACUACGUGCCAGCAGCCGCGGUAAUACGUAGGUG ..........((((..((((((......((((((((((((.....)))))))).))))......)))))))))).(((....)))..(((((((((.((....)))))...))))))... (-41.53 = -39.77 + -1.75)

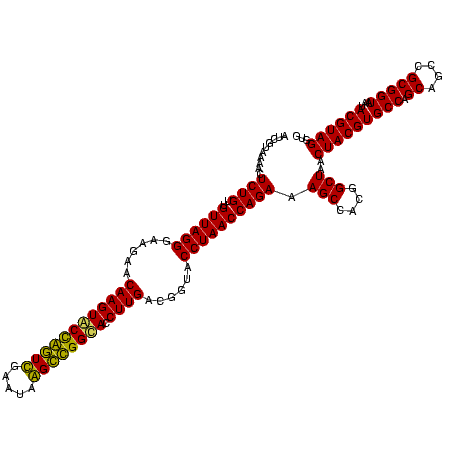
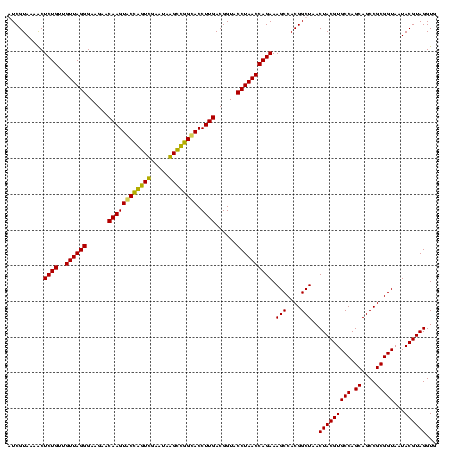
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -38.98 |
| Consensus MFE | -39.22 |
| Energy contribution | -38.79 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.03 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/400-520 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCGCCCUAUUCGAACGGUACUUGUUCUUCCCUAACAACAGAGUUUUACGAU ......((((...((((((((((((.....))).)))))))))..(((((((((((........((((((((..(.....)..)))))))).......))))))..)))))...)))).. ( -40.46) >Bsubt.0 946065 120 + 4214630/400-520 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUACCGCCCUAUUCGAACGGUACUUGUUCUUCCCUAACAACAGAGCUUUACGAU ......((((...((((((((((((.....))).)))))))))..(((((((((((........((((((((..(.....)..)))))))).......))))))..)))))...)))).. ( -40.36) >Bcere.0 604964 120 + 5224283/400-520 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGAC ......((((...((((((((((((.....))).)))))))))..(((((((((((........((((((.((.........)).)))))).......))))))..)))))...)))).. ( -37.56) >Banth.0 532469 120 + 5227293/400-520 CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCAGCUUAUUCAACUAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGAC ......((((...((((((((((((.....))).)))))))))..(((((((((((........((((((.((.........)).)))))).......))))))..)))))...)))).. ( -37.56) >consensus CACCUACGUAUUACCGCGGCUGCUGGCACGUAGUUAGCCGUGGCUUUCUGGUUAGGUACCGUCAAGGUGCCACCCUAUUCAAACAGCACUUGUUCUUCCCUAACAACAGAGUUUUACGAC ......((((...((((((((((((.....))).)))))))))..(((((((((((........(((((((((.(.....).))))))))).......))))))..)))))...)))).. (-39.22 = -38.79 + -0.44)

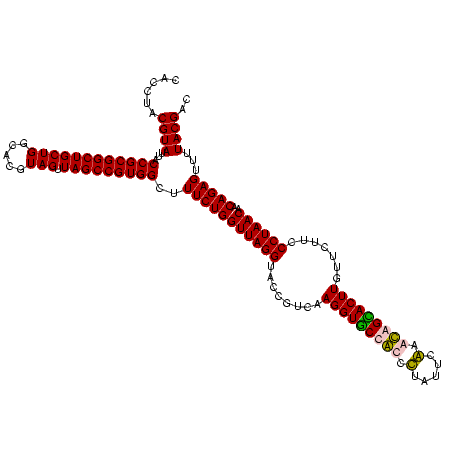
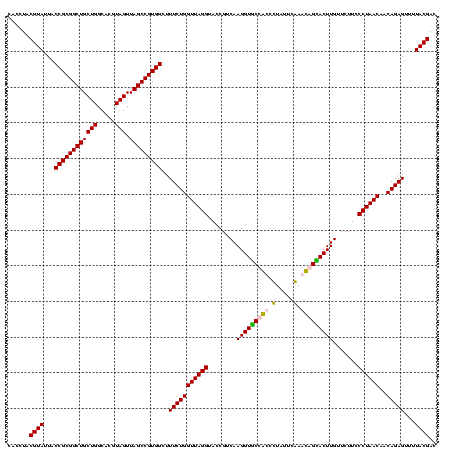
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -25.26 |
| Consensus MFE | -22.52 |
| Energy contribution | -22.27 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.560228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/560-680 CUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACCGCC ....((.((((..........((((....))))........((((.....((((.((((.((((.....((((......)))))))).))))))))......)))).....))))..)). ( -26.50) >Bsubt.0 946065 120 + 4214630/560-680 CUACGCAUUUCACCGCUACACGUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUUCCCCAGUUUCCAAUGACCCUCCCCGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACCGCC ....((.((((..........((((....))))........((((.....((((.((((.((((.....((((......)))))))).))))))))......)))).....))))..)). ( -26.50) >Bcere.0 604964 120 + 5224283/560-680 CUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCACC .......((((...((.......((((.......))))......))....((((((....(((......)))..(((((((.....)))))))..........))))))..))))..... ( -24.02) >Banth.0 532469 120 + 5227293/560-680 CUACGCAUUUCACCGCUACACAUGGAAUUCCACUUUCCUCUUCUGCACUCAAGUCUCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGUGGGCUUUCACAUCAGACUUAAGAAACCACC .......((((...((.......((((.......))))......))....((((((....(((......)))..(((((((.....)))))))..........))))))..))))..... ( -24.02) >consensus CUACGCAUUUCACCGCUACACAUGGAAUUCCACUCUCCUCUUCUGCACUCAAGUCCCCCAGUUUCCAAUGACCCUCCACGGUUGAGCCGGGGGCUUUCACAUCAGACUUAAGAAACCACC .......((((...((.......((((.......))))......))....(((((.....(((......)))..(((((((.....)))))))...........)))))..))))..... (-22.52 = -22.27 + -0.25)

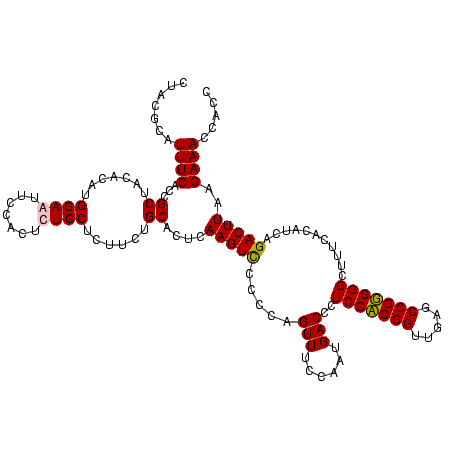
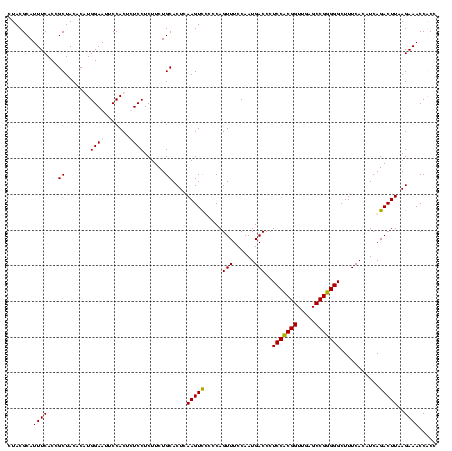
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.11 |
| Mean single sequence MFE | -36.20 |
| Consensus MFE | -36.65 |
| Energy contribution | -35.65 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.29 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/600-720 CGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGU ...((((((((.......(((((...((((.....(....)....))))(...((((((((.(.(((......)))....).))))))))...).))))).((....))))))))))... ( -37.90) >Bsubt.0 946065 120 + 4214630/600-720 CGGGGAGGGUCAUUGGAAACUGGGGAACUUGAGUGCAGAAGAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGU ...((((((((.......(((((...((((.....(....)....))))(...((((((((.(.(((......)))....).))))))))...).))))).((....))))))))))... ( -37.90) >Bcere.0 604964 120 + 5224283/600-720 CGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGU ..(.(((..((...(....)...))..))).)...(((((.......(((...((((((((.(.(((......)))....).))))))))...))).(((.((....)).)))))))).. ( -34.50) >Banth.0 532469 120 + 5227293/600-720 CGUGGAGGGUCAUUGGAAACUGGGAGACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGU ..(.(((..((...(....)...))..))).)...(((((.......(((...((((((((.(.(((......)))....).))))))))...))).(((.((....)).)))))))).. ( -34.50) >consensus CGGGGAGGGUCAUUGGAAACUGGGAAACUUGAGUGCAGAAGAGGAAAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGU ...((((((((.......(((((...((((.....(....)....))))(...((((((((.(.(((......)))....).))))))))...).))))).((....))))))))))... (-36.65 = -35.65 + -1.00)

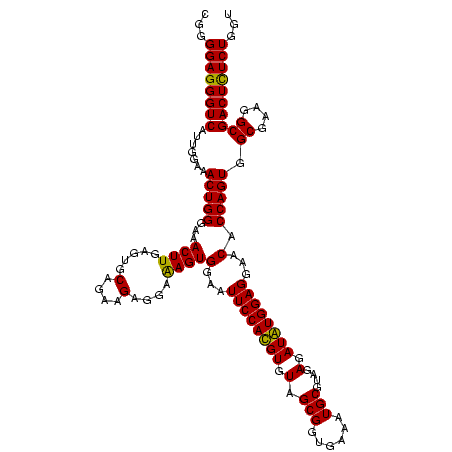
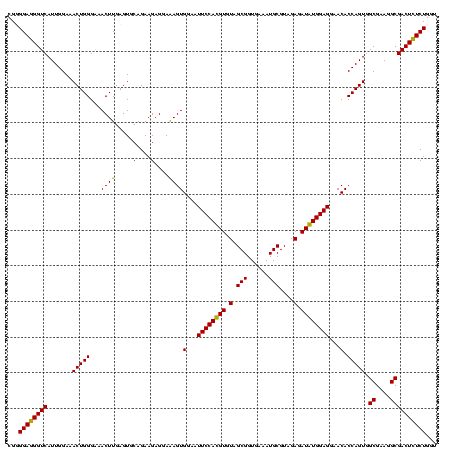
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.25 |
| Mean single sequence MFE | -39.93 |
| Consensus MFE | -37.62 |
| Energy contribution | -37.12 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.954708 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/640-760 GAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGCGCGAAAGCGUGGGGAGCGAAC ..((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).))))))).(((.......)))(((...((((....))))....))).... ( -42.60) >Bsubt.0 946065 120 + 4214630/640-760 GAGGAGAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUGUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACGCUGAGGAGCGAAAGCGUGGGGAGCGAAC ..((((((((...((((((((.(.(((......)))....).))))))))...)))..((.((....)).))))))).((.(((..((.(((((..........))))).))..))).)) ( -40.70) >Bcere.0 604964 120 + 5224283/640-760 GAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAAC .......(((...((((((((.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))........... ( -38.20) >Banth.0 532469 120 + 5227293/640-760 GAGGAAAGUGGAAUUCCAUGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUUUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAAC .......(((...((((((((.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))........... ( -38.20) >consensus GAGGAAAGUGGAAUUCCACGUGUAGCGGUGAAAUGCGUAGAGAUAUGGAGGAACACCAGUGGCGAAGGCGACUCUCUGGUCUGUAACUGACACUGAGGCGCGAAAGCGUGGGGAGCAAAC .......(((...((((((((.(.(((......)))....).))))))))...)))(((((((....))((((....)))).........)))))..((((....))))........... (-37.62 = -37.12 + -0.50)

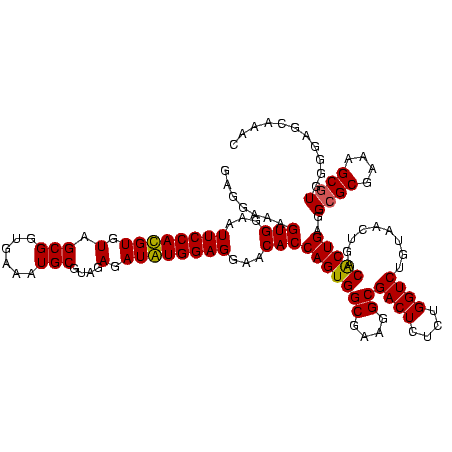
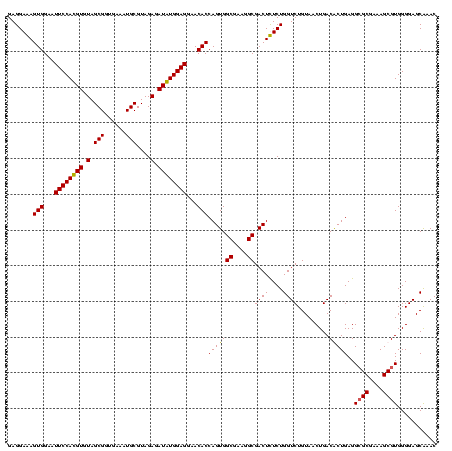
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.53 |
| Mean single sequence MFE | -50.35 |
| Consensus MFE | -51.05 |
| Energy contribution | -49.92 |
| Covariance contribution | -1.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.95 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/800-920 UGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGCAGCAAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG ((((.((..(((((((((....)))))))))..))..))))..........((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) ( -50.80) >Bsubt.0 946065 120 + 4214630/800-920 UGCUAAGUGUUAGGGGGUUUCCGCCCCUUAGUGCUGCAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG (((..((..(((((((((....)))))))))..)))))((....)).....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) ( -52.60) >Bcere.0 604964 120 + 5224283/800-920 UGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG .(((.((..(((((((((....)))))))))..))..)))...........((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) ( -49.00) >Banth.0 532469 120 + 5227293/800-920 UGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGUUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG .(((.((..(((((((((....)))))))))..))..)))...........((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) ( -49.00) >consensus UGCUAAGUGUUAGAGGGUUUCCGCCCUUUAGUGCUGAAGCUAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGG .(((.((..(((((((((....)))))))))..))..)))...........((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....)))) (-51.05 = -49.92 + -1.12)

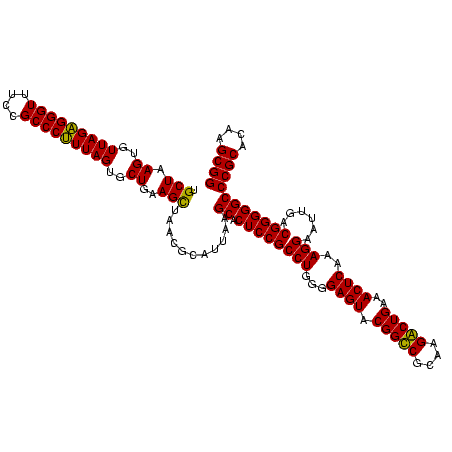
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.47 |
| Mean single sequence MFE | -40.75 |
| Consensus MFE | -41.25 |
| Energy contribution | -40.75 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.58 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.538218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/840-960 AAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. ( -39.70) >Bsubt.0 946065 120 + 4214630/840-960 UAACGCAUUAAGCACUCCGCCUGGGGAGUACGGUCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. ( -39.70) >Bcere.0 604964 120 + 5224283/840-960 UAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. ( -41.80) >Banth.0 532469 120 + 5227293/840-960 UAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGGCUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. ( -41.80) >consensus UAACGCAUUAAGCACUCCGCCUGGGGAGUACGGCCGCAAGACUGAAACUCAAAGGAAUUGACGGGGGCCCGCACAAGCGGUGGAGCAUGUGGUUUAAUUCGAAGCAACGCGAAGAACCUU ....((.....((.((((((((...((((.(((((....)))))..))))..)))......)))))))((((....))))....))....(((((..((((........))))))))).. (-41.25 = -40.75 + -0.50)

| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.92 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -37.79 |
| Energy contribution | -34.60 |
| Covariance contribution | -3.19 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.90 |
| Structure conservation index | 1.06 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590266 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/1200-1320 UACACACGUGCUACAAUGGGCAGAACAAAGGGCAGCGAAGCCGCGAGGCUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCG .................(((((((.....(((..((..((((....))))..))....))).....)))))))((((((.....(((((((........)))))))...))))))..... ( -39.30) >Bsubt.0 946065 119 + 4214630/1200-1320 UACACACGUGCUACAAUGGACAGAACAAAGGGCAGCGAA-CCGCGAGGUUAAGCCAAUCCCACAAAUCUGUUCUCAGUUCGGAUCGCAGUCUGCAACUCGACUGCGUGAAGCUGGAAUCG .................(((((((.....(((..((.((-((....))))..))....))).....)))))))((((((.....(((((((........)))))))...))))))..... ( -35.90) >Bcere.0 604964 120 + 5224283/1200-1320 UACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCG .................(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))..... ( -33.80) >Banth.0 532469 120 + 5227293/1200-1320 UACACACGUGCUACAAUGGACGGUACAAAGAGCUGCAAGACCGCGAGGUGGAGCUAAUCUCAUAAAACCGUUCUCAGUUCGGAUUGUAGGCUGCAACUCGCCUACAUGAAGCUGGAAUCG .................(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))..... ( -33.80) >consensus UACACACGUGCUACAAUGGACAGAACAAAGAGCAGCAAAACCGCGAGGUGAAGCCAAUCCCACAAAACCGUUCUCAGUUCGGAUCGCAGGCUGCAACUCGACUACAUGAAGCUGGAAUCG .................(((((((.....(((..((...(((....)))...))....))).....)))))))((((((.....(((((((........)))))))...))))))..... (-37.79 = -34.60 + -3.19)

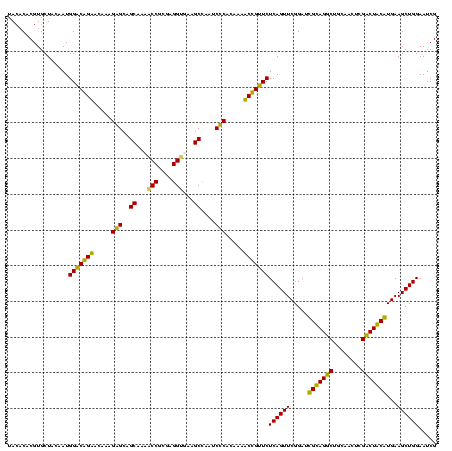
| Location | 920,518 – 920,637 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.05 |
| Mean single sequence MFE | -46.17 |
| Consensus MFE | -43.72 |
| Energy contribution | -44.98 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861307 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 119 + 4222334/1400-1520 UAACACCCGAAGUCGGUGAGGUAACCUUUU-GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU .....((((...((((((.(((..((....-)).)))...))))))...))))..........(((((((.((....))....((((((((((....))))))))))..))))))).... ( -47.80) >Bsubt.0 946065 120 + 4214630/1400-1520 UAACACCCGAAGUCGGUGAGGUAACCUUUUAGGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU .....((((...((((((.(((..((.....)).)))...))))))...))))..........(((((((.((....))....((((((((((....))))))))))..))))))).... ( -47.50) >Bcere.0 604964 119 + 5224283/1400-1520 UAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCSUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU ...((((..(.(.((((..(((..((....-)).))).))))).)..))))............(((((((.((....))....((((((((((....))))))))))..))))))).... ( -44.10) >Banth.0 532469 119 + 5227293/1400-1520 UAACACCCGAAGUCGGUGGGGUAACCUUUU-GGAGCCAGCCGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU .....((((.....((((((((..((....-)).)))..))))).....))))..........(((((((.((....))....((((((((((....))))))))))..))))))).... ( -45.30) >consensus UAACACCCGAAGUCGGUGAGGUAACCUUUU_GGAGCCAGCCGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUU .....((((...((((((((((..((.....)).)))..)))))))...))))..........(((((((.((....))....((((((((((....))))))))))..))))))).... (-43.72 = -44.98 + 1.25)

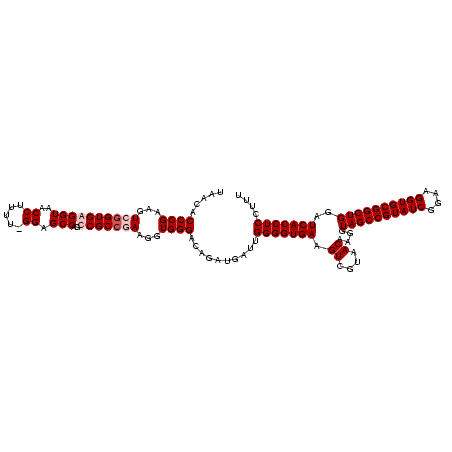
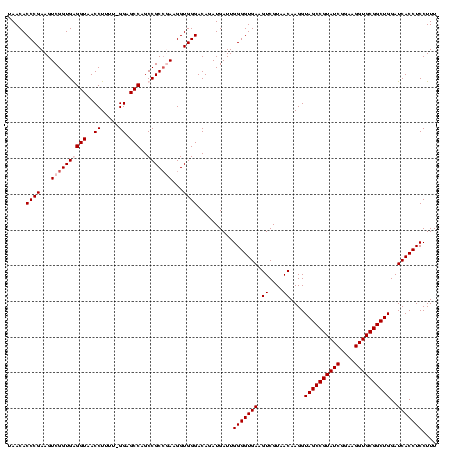
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -43.62 |
| Consensus MFE | -32.38 |
| Energy contribution | -32.88 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.74 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987222 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/1440-1560 CGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACCUUGCGGUCUUAU ..(((..(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....)))).......)))...((((((((....)))))))) ( -45.10) >Bsubt.0 946065 119 + 4214630/1440-1560 CGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACCUUG-GGUCUUAU ..(((..(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))((((.....)))).......)))...((((((((...-)))))))) ( -43.60) >Bcere.0 604964 114 + 5224283/1440-1560 CGCSUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAU .......(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))(((.......)))..(((((((------(((...)))))))))).. ( -41.80) >Banth.0 532469 114 + 5227293/1440-1560 CGCCUAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAU ((((...))))...((...((..(((((((.((....))....((((((((((....))))))))))..))))).))..))..))....(((((((------(((...)))))))))).. ( -44.00) >consensus CGCCGAAGGUGGGACAGAUGAUUGGGGUGAAGUCGUAACAAGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAAUAGACG______AACACCGUGCAGCAAUAU .......(((((..(..((((((.......))))))........(((((((((....))))))))))..)))))(((.......)))...............((((((....)))))).. (-32.38 = -32.88 + 0.50)

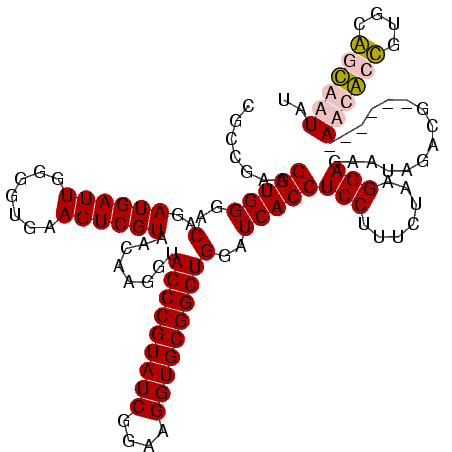
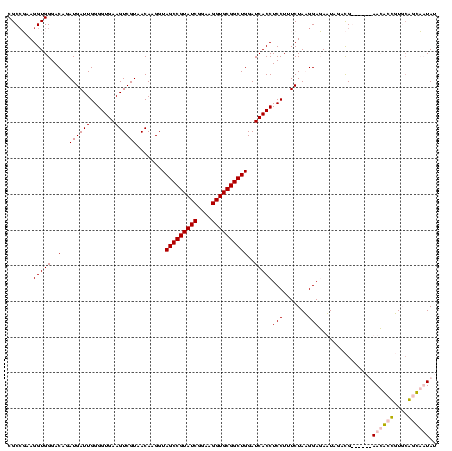
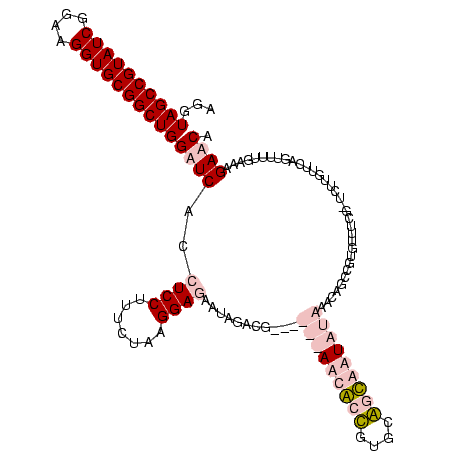
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.88 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -24.58 |
| Energy contribution | -27.57 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.63 |
| Structure conservation index | 0.59 |
| SVM decision value | 4.42 |
| SVM RNA-class probability | 0.999893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/1480-1600 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACCUUGCGGUCUUAUAGACAGGUGCGUUUGGAUCUUGUUUAGUUUUGAAGGAUCA ...((((((((((....))))))))))((((...((((.....)))).......((((((.(((((((....)))))))((((((((..(.....)..))))))))))))))...)))). ( -43.40) >Bsubt.0 946065 119 + 4214630/1480-1600 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACCUUG-GGUCUUAUAAACAGAACGUUCCCUGUCUUGUUUAGUUUUGAAGGAUCA ...((((((((((....))))))))))((((...((((.....)))).......((((((.(((((((...-)))))))((((((..(((.....)))..))))))))))))...)))). ( -41.40) >Bcere.0 604964 113 + 5224283/1480-1600 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUCG-UUUUGUUCAGUUUUGAGAGAACU ....(((((((((....)))))))))(((.(((((((.......)))).(((((((------(((...))))))))))..........))).)))(-((((.(((......)))))))). ( -40.60) >Banth.0 532469 113 + 5227293/1480-1600 AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUCG-UUUUGUUCAGUUUUGAGAGAACU ....(((((((((....)))))))))(((.(((((((.......)))).(((((((------(((...))))))))))..........))).)))(-((((.(((......)))))))). ( -40.60) >consensus AGGUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAAUAGACG______AACACCGUGCAGCAAUAUAAACAGCCGUGUUUCG_UCUUGUUCAGUUUUGAAAGAACA ...((((((((((....))))))))))((((..((((.......))))...........(((((((((....)))))))))..................................)))). (-24.58 = -27.57 + 3.00)

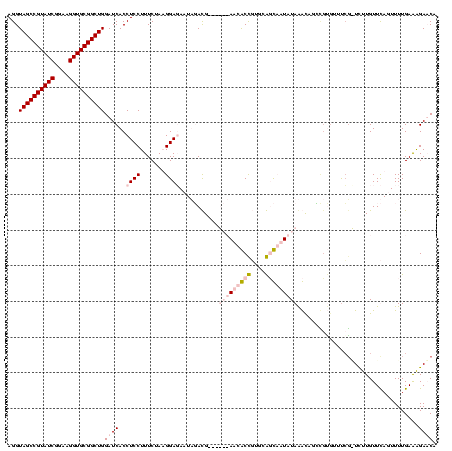
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.88 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/1480-1600 UGAUCCUUCAAAACUAAACAAGAUCCAAACGCACCUGUCUAUAAGACCGCAAGGUCUUAUAUUCCGUAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCU .((((................))))......((((....(((((((((....)))))))))............((((.....))))))))....(((((............))))).... ( -29.99) >Bsubt.0 946065 119 + 4214630/1480-1600 UGAUCCUUCAAAACUAAACAAGACAGGGAACGUUCUGUUUAUAAGACC-CAAGGUCUUAUAUUCCGUAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCU .((((((((....((((....(((((((....)))))))(((((((((-...)))))))))...............))))....)))).)))).(((((............))))).... ( -29.80) >Bcere.0 604964 113 + 5224283/1480-1600 AGUUCUCUCAAAACUGAACAAAA-CGAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CAUCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCU .((((..........))))....-........(((.......((((((((((...)))------)))))))(((((.......)))))...)))(((((............))))).... ( -27.20) >Banth.0 532469 113 + 5227293/1480-1600 AGUUCUCUCAAAACUGAACAAAA-CGAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CAUCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCU .((((..........))))....-........(((.......((((((((((...)))------)))))))(((((.......)))))...)))(((((............))))).... ( -27.20) >consensus AGAUCCCUCAAAACUAAACAAAA_CGAAACACGGAAACUUAUAAGAACGAAAAGCCUU______CAUAAAAUAUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUACCU .((((((((....(((................(....).(((((((((....)))))))))................)))....)))).)))).(((((............))))).... (-15.10 = -15.85 + 0.75)

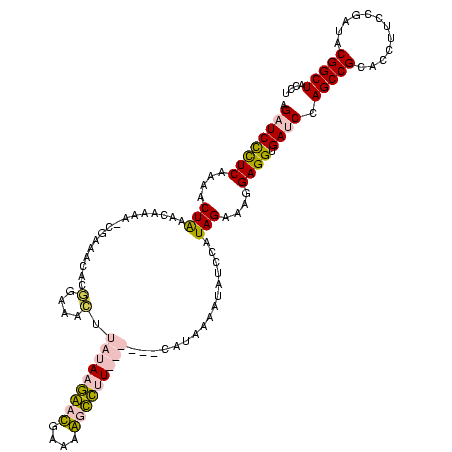
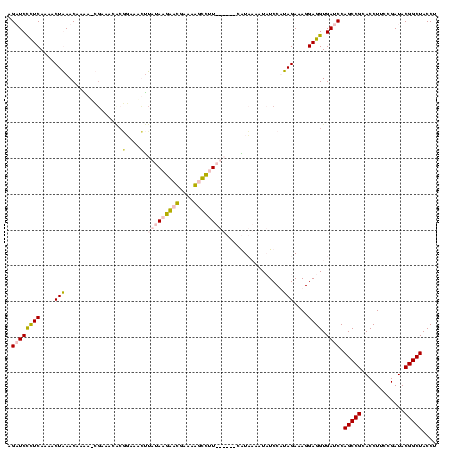
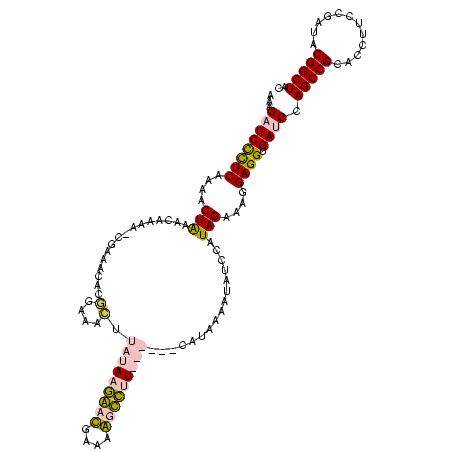
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -24.58 |
| Energy contribution | -27.57 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.86 |
| Structure conservation index | 0.59 |
| SVM decision value | 4.73 |
| SVM RNA-class probability | 0.999943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/1482-1602 GUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACCUUGCGGUCUUAUAGACAGGUGCGUUUGGAUCUUGUUUAGUUUUGAAGGAUCAUU .((((((((((....))))))))))((((...((((.....)))).......((((((.(((((((....)))))))((((((((..(.....)..))))))))))))))...))))... ( -43.40) >Bsubt.0 946065 119 + 4214630/1482-1602 GUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAUAUUAUACGGAAUAUAAGACCUUG-GGUCUUAUAAACAGAACGUUCCCUGUCUUGUUUAGUUUUGAAGGAUCAUU .((((((((((....))))))))))((((...((((.....)))).......((((((.(((((((...-)))))))((((((..(((.....)))..))))))))))))...))))... ( -41.40) >Bcere.0 604964 113 + 5224283/1482-1602 GUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUCG-UUUUGUUCAGUUUUGAGAGAACUAU ..(((((((((....)))))))))(((.(((((((.......)))).(((((((------(((...))))))))))..........))).)))(-((((.(((......))))))))... ( -40.60) >Banth.0 532469 113 + 5227293/1482-1602 GUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAUGGAGAAUUGAUG------AACGCUGUUCAUCAAUAUAAGUUUCCGUGUUUCG-UUUUGUUCAGUUUUGAGAGAACUAU ..(((((((((....)))))))))(((.(((((((.......)))).(((((((------(((...))))))))))..........))).)))(-((((.(((......))))))))... ( -40.60) >consensus GUAGCCGUAUCGGAAGGUGCGGCUGGAUCACCUCCUUUCUAAGGAGAAUAGACG______AACACCGUGCAGCAAUAUAAACAGCCGUGUUUCG_UCUUGUUCAGUUUUGAAAGAACAAU .((((((((((....))))))))))((((..((((.......))))...........(((((((((....)))))))))..................................))))... (-24.58 = -27.57 + 3.00)

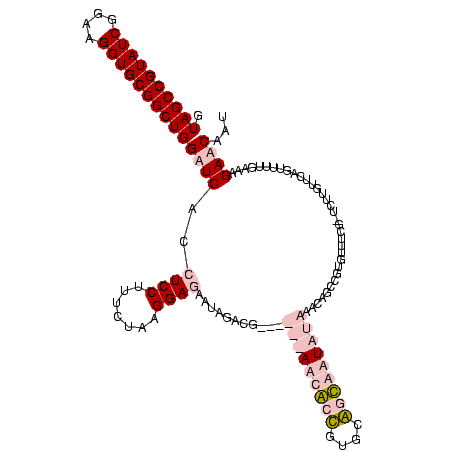
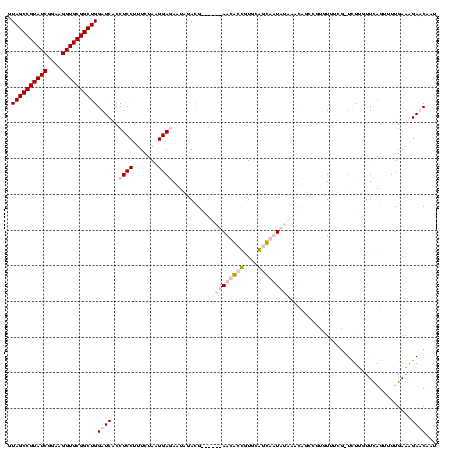
| Location | 920,518 – 920,638 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.32 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -15.10 |
| Energy contribution | -15.85 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.47 |
| Structure conservation index | 0.53 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.969006 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 920518 120 + 4222334/1482-1602 AAUGAUCCUUCAAAACUAAACAAGAUCCAAACGCACCUGUCUAUAAGACCGCAAGGUCUUAUAUUCCGUAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUAC ...((((................))))......((((....(((((((((....)))))))))............((((.....))))))))....(((((............))))).. ( -29.99) >Bsubt.0 946065 119 + 4214630/1482-1602 AAUGAUCCUUCAAAACUAAACAAGACAGGGAACGUUCUGUUUAUAAGACC-CAAGGUCUUAUAUUCCGUAUAAUAUCCUUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUAC ...((((((((....((((....(((((((....)))))))(((((((((-...)))))))))...............))))....)))).)))).(((((............))))).. ( -29.80) >Bcere.0 604964 113 + 5224283/1482-1602 AUAGUUCUCUCAAAACUGAACAAAA-CGAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CAUCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUAC ...((((..........))))....-........(((.......((((((((((...)))------)))))))(((((.......)))))...)))(((((............))))).. ( -27.20) >Banth.0 532469 113 + 5227293/1482-1602 AUAGUUCUCUCAAAACUGAACAAAA-CGAAACACGGAAACUUAUAUUGAUGAACAGCGUU------CAUCAAUUCUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUAC ...((((..........))))....-........(((.......((((((((((...)))------)))))))(((((.......)))))...)))(((((............))))).. ( -27.20) >consensus AAAGAUCCCUCAAAACUAAACAAAA_CGAAACACGGAAACUUAUAAGAACGAAAAGCCUU______CAUAAAAUAUCCAUAGAAAGGAGGUGAUCCAGCCGCACCUUCCGAUACGGCUAC ...((((((((....(((................(....).(((((((((....)))))))))................)))....)))).)))).(((((............))))).. (-15.10 = -15.85 + 0.75)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:24:43 2006