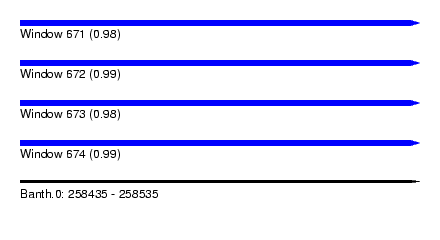
| Sequence ID | Banth.0 |
|---|---|
| Location | 258,435 – 258,535 |
| Length | 100 |
| Max. P | 0.993984 |

| Location | 258,435 – 258,535 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 67.71 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
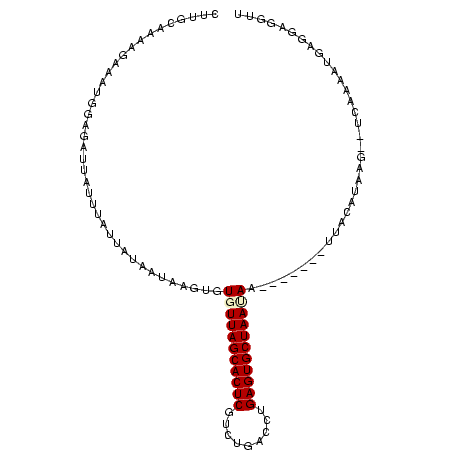
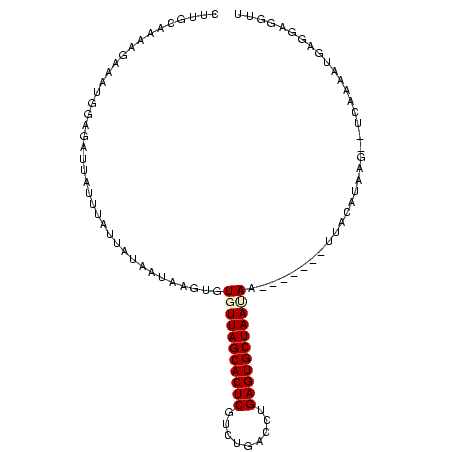
>Banth.0 258435 100 + 5227293 CUUGCAAAAGAAAUUGAGAUUAUUUAUUAUUAUAAGUGUGUUAGCACUCGGGUGACUUGAGUGCUAAUAAAUAAAAUUUACAUAA----CAAAAUGAGGAGGUU (((.(.....(((((.....(((((((....)))))))(((((((((((((....).)))))))))))).....))))).(((..----....))).).))).. ( -22.50) >Bcere.0 292306 98 + 5224283 CUUGCAAAAGAAAUUGAGAUUAUUUAUUAUUAUAAGUGUGUUAGCACUCGAGUGACUUGAGUGCUAAUAAAAAAA--UUACAUAA----CAAAAUGAGGAGGUU (((.(...............(((((((....)))))))((((((((((((((...))))))))))))))......--........----........).))).. ( -21.90) >Bhalo.0 594652 102 + 4202352 CUUGCAAAAUAAGCAGGUUCUGAUUAUCAUUGUAACUGUGUUAGCACUCGUUUGAAGAGAGUGCUAACAAGGAAA-AUCUUAUAAG-CUCUAAAGGAGGGUGUU .((((((.((((.(((...))).))))..))))))((.(((((((((((.........))))))))))).))...-.........(-((((.....)))))... ( -26.90) >Bclau.0 956367 99 + 4303871 CUUGCAAAACAAGUGGGGAAUGAUUAUUAUAAUAAAUGUGUUAGCACUCAUCGAAUCCGAGUGCUAAUAAG-----ACUACAUACGUCUUCAAAGGAGGGUGUU ............((((.......((((....))))...(((((((((((.........)))))))))))..-----.))))..((..((((....))))..)). ( -22.80) >Blich.0 626514 95 + 4222334 CUUGAAAUUGCCUGGGAGAUUCUUUAUUAUAAGAAUUGUGUUAGCACUCACCGAGUGCGAGUGCUAAAA--------UUACAUAUGAAAA-UAUUGAGGAGGUU .....(((..(((.((.(((((((......)))))))((((((((((((.(.....).)))))))))..--------.))).........-..)).)))..))) ( -22.10) >Bsubt.0 649347 96 + 4214630 CUUGAAAUUGGAAGGGAGAUUCUUUAUUAUAAGAAUUGUGUUAGCACUCUUUAGUGCUGAGUGCUAAAA--------UUACAUAUUCAUACUAUUGAGGAGGUU ...................(((((((.((((.(((((((((((((((((.........)))))))))..--------.)))).)))).)).)).)))))))... ( -20.90) >consensus CUUGCAAAAGAAAUGGAGAUUAUUUAUUAUAAUAAGUGUGUUAGCACUCGUCUGACCUGAGUGCUAAUAA_______UUACAUAAG__UCAAAAUGAGGAGGUU ......................................(((((((((((.........)))))))))))................................... (-13.57 = -13.82 + 0.25)


| Location | 258,435 – 258,535 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 67.71 |
| Mean single sequence MFE | -17.96 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
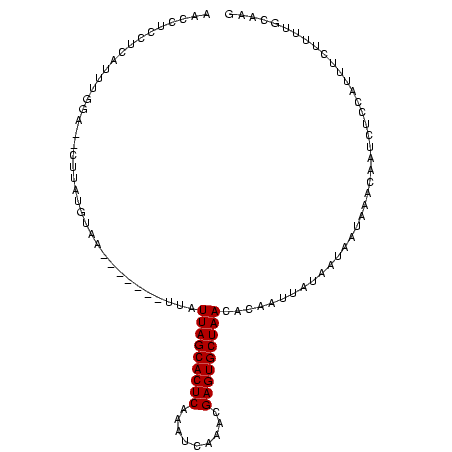
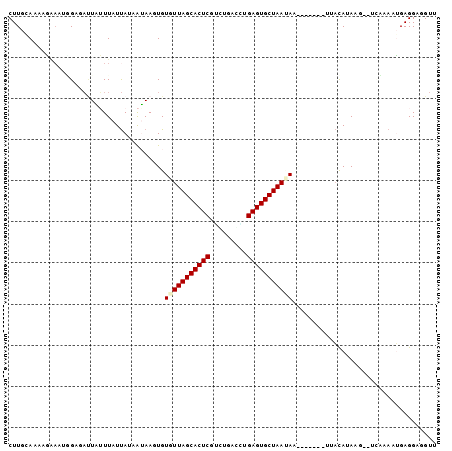
>Banth.0 258435 100 + 5227293 AACCUCCUCAUUUUG----UUAUGUAAAUUUUAUUUAUUAGCACUCAAGUCACCCGAGUGCUAACACACUUAUAAUAAUAAAUAAUCUCAAUUUCUUUUGCAAG ...............----...((((((.........(((((((((.........))))))))).....((((....))))...............)))))).. ( -14.20) >Bcere.0 292306 98 + 5224283 AACCUCCUCAUUUUG----UUAUGUAA--UUUUUUUAUUAGCACUCAAGUCACUCGAGUGCUAACACACUUAUAAUAAUAAAUAAUCUCAAUUUCUUUUGCAAG ...........((((----((((....--........(((((((((.((...)).)))))))))..........))))))))...................... ( -14.95) >Bhalo.0 594652 102 + 4202352 AACACCCUCCUUUAGAG-CUUAUAAGAU-UUUCCUUGUUAGCACUCUCUUCAAACGAGUGCUAACACAGUUACAAUGAUAAUCAGAACCUGCUUAUUUUGCAAG ................(-(..(((((..-......(((((((((((.........)))))))))))(((......((.....))....))))))))...))... ( -22.40) >Bclau.0 956367 99 + 4303871 AACACCCUCCUUUGAAGACGUAUGUAGU-----CUUAUUAGCACUCGGAUUCGAUGAGUGCUAACACAUUUAUUAUAAUAAUCAUUCCCCACUUGUUUUGCAAG ..............(((((.......))-----))).((((((((((.......))))))))))...........................((((.....)))) ( -19.90) >Blich.0 626514 95 + 4222334 AACCUCCUCAAUA-UUUUCAUAUGUAA--------UUUUAGCACUCGCACUCGGUGAGUGCUAACACAAUUCUUAUAAUAAAGAAUCUCCCAGGCAAUUUCAAG ..(((........-.............--------..(((((((((((.....)))))))))))....((((((......)))))).....))).......... ( -20.40) >Bsubt.0 649347 96 + 4214630 AACCUCCUCAAUAGUAUGAAUAUGUAA--------UUUUAGCACUCAGCACUAAAGAGUGCUAACACAAUUCUUAUAAUAAAGAAUCUCCCUUCCAAUUUCAAG ................((((..((...--------..(((((((((.........)))))))))....((((((......))))))........))..)))).. ( -15.90) >consensus AACCUCCUCAUUUGGA__CUUAUGUAA_______UUAUUAGCACUCAAAUCAAACGAGUGCUAACACAAUUAUAAUAAUAAACAAUCUCCAUUUCUUUUGCAAG .....................................(((((((((.........)))))))))........................................ (-11.70 = -11.70 + 0.00)

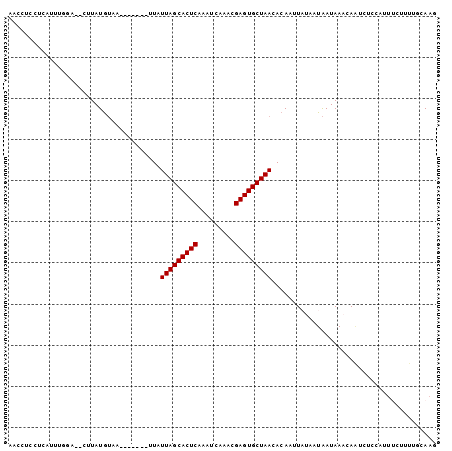
| Location | 258,435 – 258,535 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 67.71 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -13.57 |
| Energy contribution | -13.82 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
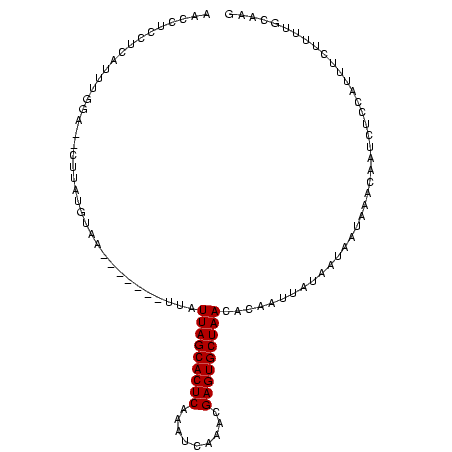
>Banth.0 258435 100 + 5227293/0-104 CUUGCAAAAGAAAUUGAGAUUAUUUAUUAUUAUAAGUGUGUUAGCACUCGGGUGACUUGAGUGCUAAUAAAUAAAAUUUACAUAA----CAAAAUGAGGAGGUU (((.(.....(((((.....(((((((....)))))))(((((((((((((....).)))))))))))).....))))).(((..----....))).).))).. ( -22.50) >Bcere.0 292306 98 + 5224283/0-104 CUUGCAAAAGAAAUUGAGAUUAUUUAUUAUUAUAAGUGUGUUAGCACUCGAGUGACUUGAGUGCUAAUAAAAAAA--UUACAUAA----CAAAAUGAGGAGGUU (((.(...............(((((((....)))))))((((((((((((((...))))))))))))))......--........----........).))).. ( -21.90) >Bhalo.0 594652 102 + 4202352/0-104 CUUGCAAAAUAAGCAGGUUCUGAUUAUCAUUGUAACUGUGUUAGCACUCGUUUGAAGAGAGUGCUAACAAGGAAA-AUCUUAUAAG-CUCUAAAGGAGGGUGUU .((((((.((((.(((...))).))))..))))))((.(((((((((((.........))))))))))).))...-.........(-((((.....)))))... ( -26.90) >Bclau.0 956367 99 + 4303871/0-104 CUUGCAAAACAAGUGGGGAAUGAUUAUUAUAAUAAAUGUGUUAGCACUCAUCGAAUCCGAGUGCUAAUAAG-----ACUACAUACGUCUUCAAAGGAGGGUGUU ............((((.......((((....))))...(((((((((((.........)))))))))))..-----.))))..((..((((....))))..)). ( -22.80) >Blich.0 626514 95 + 4222334/0-104 CUUGAAAUUGCCUGGGAGAUUCUUUAUUAUAAGAAUUGUGUUAGCACUCACCGAGUGCGAGUGCUAAAA--------UUACAUAUGAAAA-UAUUGAGGAGGUU .....(((..(((.((.(((((((......)))))))((((((((((((.(.....).)))))))))..--------.))).........-..)).)))..))) ( -22.10) >Bsubt.0 649347 96 + 4214630/0-104 CUUGAAAUUGGAAGGGAGAUUCUUUAUUAUAAGAAUUGUGUUAGCACUCUUUAGUGCUGAGUGCUAAAA--------UUACAUAUUCAUACUAUUGAGGAGGUU ...................(((((((.((((.(((((((((((((((((.........)))))))))..--------.)))).)))).)).)).)))))))... ( -20.90) >consensus CUUGCAAAAGAAAUGGAGAUUAUUUAUUAUAAUAAGUGUGUUAGCACUCGUCUGACCUGAGUGCUAAUAA_______UUACAUAAG__UCAAAAUGAGGAGGUU ......................................(((((((((((.........)))))))))))................................... (-13.57 = -13.82 + 0.25)

| Location | 258,435 – 258,535 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 67.71 |
| Mean single sequence MFE | -17.96 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.70 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.44 |
| SVM RNA-class probability | 0.993984 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
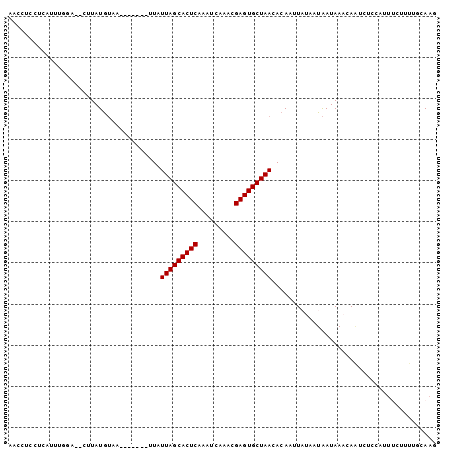
>Banth.0 258435 100 + 5227293/0-104 AACCUCCUCAUUUUG----UUAUGUAAAUUUUAUUUAUUAGCACUCAAGUCACCCGAGUGCUAACACACUUAUAAUAAUAAAUAAUCUCAAUUUCUUUUGCAAG ...............----...((((((.........(((((((((.........))))))))).....((((....))))...............)))))).. ( -14.20) >Bcere.0 292306 98 + 5224283/0-104 AACCUCCUCAUUUUG----UUAUGUAA--UUUUUUUAUUAGCACUCAAGUCACUCGAGUGCUAACACACUUAUAAUAAUAAAUAAUCUCAAUUUCUUUUGCAAG ...........((((----((((....--........(((((((((.((...)).)))))))))..........))))))))...................... ( -14.95) >Bhalo.0 594652 102 + 4202352/0-104 AACACCCUCCUUUAGAG-CUUAUAAGAU-UUUCCUUGUUAGCACUCUCUUCAAACGAGUGCUAACACAGUUACAAUGAUAAUCAGAACCUGCUUAUUUUGCAAG ................(-(..(((((..-......(((((((((((.........)))))))))))(((......((.....))....))))))))...))... ( -22.40) >Bclau.0 956367 99 + 4303871/0-104 AACACCCUCCUUUGAAGACGUAUGUAGU-----CUUAUUAGCACUCGGAUUCGAUGAGUGCUAACACAUUUAUUAUAAUAAUCAUUCCCCACUUGUUUUGCAAG ..............(((((.......))-----))).((((((((((.......))))))))))...........................((((.....)))) ( -19.90) >Blich.0 626514 95 + 4222334/0-104 AACCUCCUCAAUA-UUUUCAUAUGUAA--------UUUUAGCACUCGCACUCGGUGAGUGCUAACACAAUUCUUAUAAUAAAGAAUCUCCCAGGCAAUUUCAAG ..(((........-.............--------..(((((((((((.....)))))))))))....((((((......)))))).....))).......... ( -20.40) >Bsubt.0 649347 96 + 4214630/0-104 AACCUCCUCAAUAGUAUGAAUAUGUAA--------UUUUAGCACUCAGCACUAAAGAGUGCUAACACAAUUCUUAUAAUAAAGAAUCUCCCUUCCAAUUUCAAG ................((((..((...--------..(((((((((.........)))))))))....((((((......))))))........))..)))).. ( -15.90) >consensus AACCUCCUCAUUUGGA__CUUAUGUAA_______UUAUUAGCACUCAAAUCAAACGAGUGCUAACACAAUUAUAAUAAUAAACAAUCUCCAUUUCUUUUGCAAG .....................................(((((((((.........)))))))))........................................ (-11.70 = -11.70 + 0.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:24:25 2006