
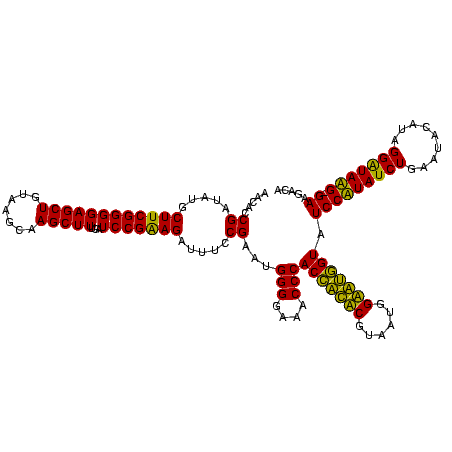
| Sequence ID | Banth.0 |
|---|---|
| Location | 247,889 – 248,009 |
| Length | 120 |
| Max. P | 0.999301 |

| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -41.80 |
| Consensus MFE | -42.35 |
| Energy contribution | -39.60 |
| Covariance contribution | -2.75 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.50 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.999301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/80-200 AACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACA .....((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........)))))))-))..... ( -42.20) >Bcere.0 281279 119 + 5224283/80-200 AACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACA .....((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........)))))))-))..... ( -42.20) >Bsubt.0 636660 120 + 4214630/80-200 AACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCA ..........((((((((....))).))))).(((((....(((......)))...(((....)))(((((((......)))))))...(((((((.........)))))))..))))). ( -41.40) >Blich.0 613689 120 + 4222334/80-200 AACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCA ..........((((((((....))).))))).(((((....(((......)))...(((....)))(((((((......)))))))...(((((((.........)))))))..))))). ( -41.40) >consensus AACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCACACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAAG_GAAGACA .....((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........))))))).))..... (-42.35 = -39.60 + -2.75)


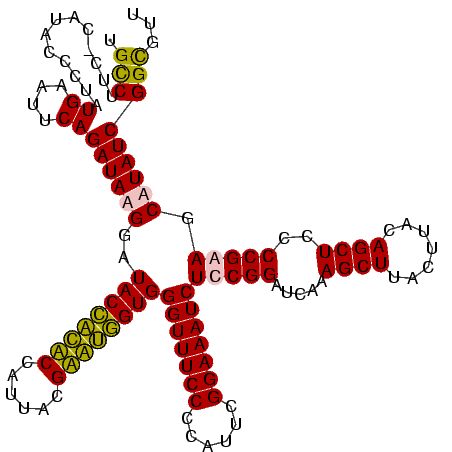
| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -31.95 |
| Consensus MFE | -30.40 |
| Energy contribution | -29.15 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.965421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/80-200 UGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUU .(((.((-((((..((........)).))))))((((((((......))))))))(((((((......)))))))(((((.....((((.......))))..))))).......)))... ( -29.60) >Bcere.0 281279 119 + 5224283/80-200 UGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUU .(((.((-((((..((........)).))))))((((((((......))))))))(((((((......)))))))(((((.....((((.......))))..))))).......)))... ( -29.60) >Bsubt.0 636660 120 + 4214630/80-200 UGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUU .(((...(((((...))))).....((((((...(((((((......)))))))(((.......(((((((....)))))))...((((.......)))).)))....)))))))))... ( -34.30) >Blich.0 613689 120 + 4222334/80-200 UGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUU .(((...(((((...))))).....((((((...(((((((......)))))))(((.......(((((((....)))))))...((((.......)))).)))....)))))))))... ( -34.30) >consensus UGCCUUC_CAUACCCUAUGAAUUCAGAUAAGGAUACCACACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUU .(((.............((....))((((((..((((((((......))))))))(((((((......)))))))(((((.....((((.......))))..))))).)))))))))... (-30.40 = -29.15 + -1.25)

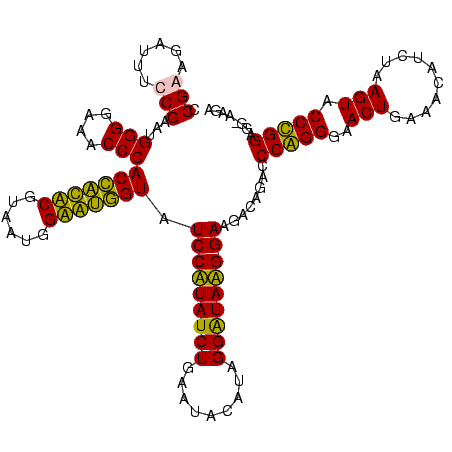
| Location | 247,889 – 248,007 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -42.25 |
| Consensus MFE | -38.60 |
| Energy contribution | -36.10 |
| Covariance contribution | -2.50 |
| Combinations/Pair | 1.23 |
| Mean z-score | -4.19 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 118 + 5227293/120-240 CCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGG-AAGA .......(((((....(((....)))(((((((......))))))).(((((((((.........)))))))-))........(((((..(((..........))).))))).))-))). ( -40.90) >Bcere.0 281279 118 + 5224283/120-240 CCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGG-AAGA .......(((((....(((....)))(((((((......))))))).(((((((((.........)))))))-))........(((((..(((..........))).))))).))-))). ( -40.90) >Bsubt.0 636660 119 + 4214630/120-240 CCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGG-AAGA .(((......)))...(((....)))(((((((......))))))).(((...((((..(((.((.....)).)))..)))).(((((..(((..........))).))))).))-)... ( -43.70) >Blich.0 613689 120 + 4222334/120-240 CCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGGAAGA .(((......)))...(((....)))(((((((......))))))).(((...((((..(((.((.....)).)))..)))).(((((..(((..........))).)))))..)))... ( -43.50) >consensus CCGAAGAUUUCCGAAUGGGGAAACCCACCACACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAAG_GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCCGGAGG_AAGA .(((......)))...(((....)))(((((((......))))))).(((((((((.........))))))).))........(((((..(((..........))).)))))........ (-38.60 = -36.10 + -2.50)

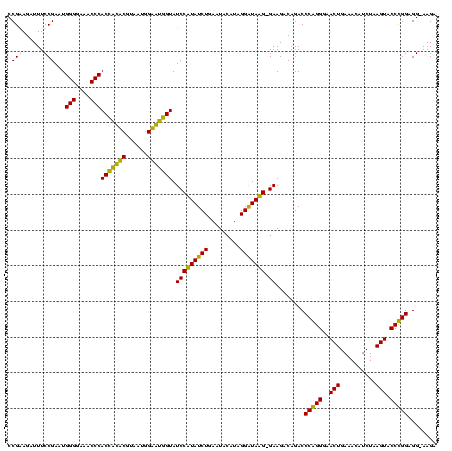
| Location | 247,889 – 248,007 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.64 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -33.90 |
| Energy contribution | -31.90 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.76 |
| SVM RNA-class probability | 0.996888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 118 + 5227293/120-240 UCUU-CCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGG ...(-((((((((.(((..((....)))))..)))))(((((.....-..(((.....)))..))))).))))((((((((......))))))))(((((((......)))))))..... ( -31.50) >Bcere.0 281279 118 + 5224283/120-240 UCUU-CCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGG ...(-((((((((.(((..((....)))))..)))))(((((.....-..(((.....)))..))))).))))((((((((......))))))))(((((((......)))))))..... ( -31.50) >Bsubt.0 636660 119 + 4214630/120-240 UCUU-CCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGG ....-...(((((.(((..((....)))))..)))))(((((..(((...........)))..))))).((((((((((((......))))))))(((((((......))))))))))). ( -37.60) >Blich.0 613689 120 + 4222334/120-240 UCUUCCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGG ........(((((.(((..((....)))))..)))))(((((..(((...........)))..))))).((((((((((((......))))))))(((((((......))))))))))). ( -37.60) >consensus UCUU_CCUCCAGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUC_CAUACCCUAUGAAUUCAGAUAAGGAUACCACACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGG ........(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..(((((((((((......))))))))(((((((......)))))))))).. (-33.90 = -31.90 + -2.00)

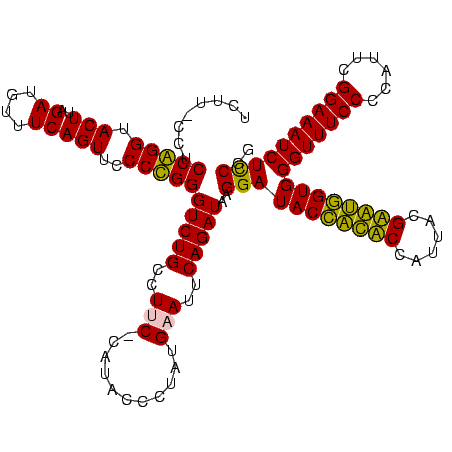
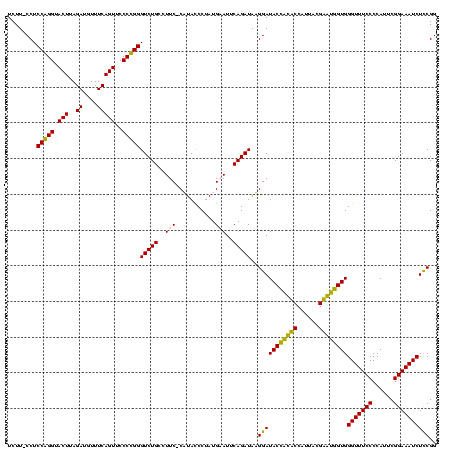
| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.08 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -34.74 |
| Energy contribution | -34.05 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/280-400 GAACAUAGCCCAAACCAAGAGG-CUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCC ..............((((((((-....))))))))((((((...((.((((((.....(((.......)))......((((....(((((....))))))))))))))).)).)))))). ( -40.40) >Bcere.0 281279 119 + 5224283/280-400 GAACAUAGCCCAAACCAAGAGG-CUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCC ..............((((((((-....))))))))((((((...((.((((((.....(((.......)))......((((....(((((....))))))))))))))).)).)))))). ( -40.40) >Bsubt.0 636660 118 + 4214630/280-400 GGA-UUAGCCCAAACCAAGAGG-CUUGCCUCUUGGGGUUGUAGGACACUCUGUACGGAGUUACAAAGGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGCCC ((.-.....))...((((((((-....))))))))((((((...((.((((.(((...(((.......)))...))).(((..(.(((((....))))))..))).)))))).)))))). ( -38.80) >Blich.0 613689 119 + 4222334/280-400 GGA-ACAGCCCAAACCAAGAGGGCUUGCCUCUUGGGGUUGUAGGACACUCUGUACGGAGUUGCAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGCCC ((.-.....)).........((((((((((((((.(((((((....(((((....)))))))))...................(.(((((....))))))))).)))))))))..))))) ( -38.60) >consensus GAA_AUAGCCCAAACCAAGAGG_CUUGCCUCUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGAGACCUGGAAAGGCCCGCCAUAGAAGGUAACAACCC ..............((((((((.....))))))))((((((...((..(((((.((...((((...........)))).))..(.(((((....))))))...)))))..)).)))))). (-34.74 = -34.05 + -0.69)

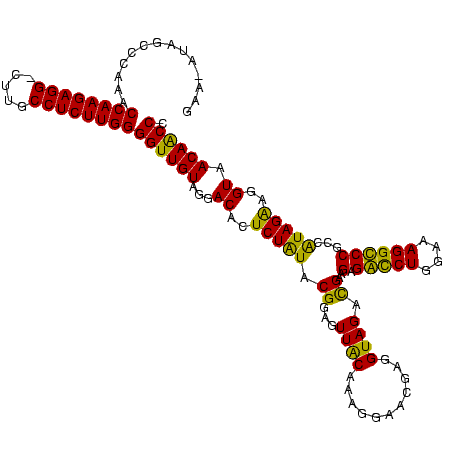
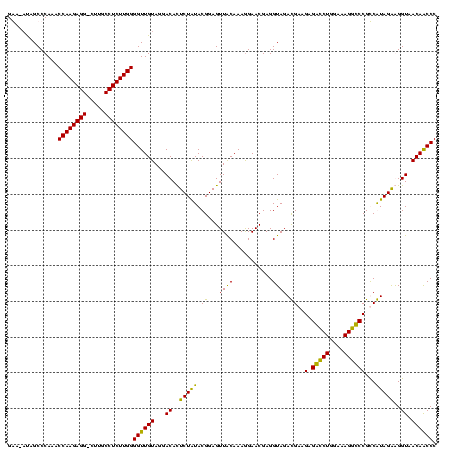
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -40.66 |
| Consensus MFE | -39.25 |
| Energy contribution | -38.56 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/320-440 UAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCCCGUAGUCGAAACUUCGUUCUCUCUUGAAUGUAUCCUGAGU ((((((((((.....((((.......((((((((((.(((((.(.(((((....)))))).))...(.((((....)))))...)))...)))))))))))))).)))))..)))))... ( -41.91) >Bcere.0 281279 120 + 5224283/320-440 UAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCCCGUAGUCGAAACUUCGUUCUCUCUUGAAUGUAUCCUGAGU ((((((((((.....((((.......((((((((((.(((((.(.(((((....)))))).))...(.((((....)))))...)))...)))))))))))))).)))))..)))))... ( -41.91) >Bsubt.0 636660 120 + 4214630/320-440 UAGGACACUCUGUACGGAGUUACAAAGGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGCCCUGUAGUCAAAACUUCGUUCUCUCCUGAGUGGAUCCUGAGU ((((((((((.....((((.......((((((((((...(((.(.(((((....))))))...(((((..........)))))..)))..)))))))))))))).)))))..)))))... ( -39.41) >Blich.0 613689 120 + 4222334/320-440 UAGGACACUCUGUACGGAGUUGCAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGCCCUGUAGUCAAAACUUCGUUCUCUCCUGAGUGGAUCCUGAGU ((((((((((.....((((.......((((((((((...(((.(.(((((....))))))...(((((..........)))))..)))..)))))))))))))).)))))..)))))... ( -39.41) >consensus UAGGACACUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGAGACCUGGAAAGGCCCGCCAUAGAAGGUAACAACCCCGUAGUCAAAACUUCGUUCUCUCCUGAAUGGAUCCUGAGU ((((((((((.....((((.......((((((((((..((((.(.(((((....)))))).))))...(((.......))).........)))))))))))))).)))))..)))))... (-39.25 = -38.56 + -0.69)

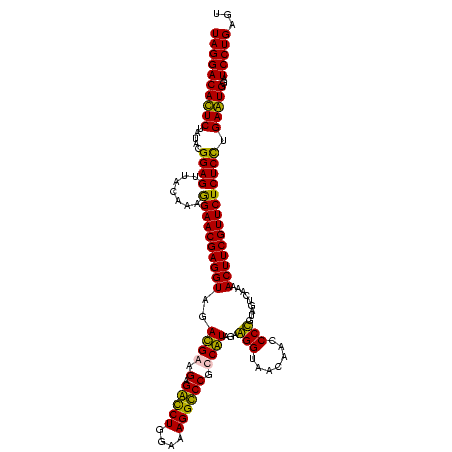
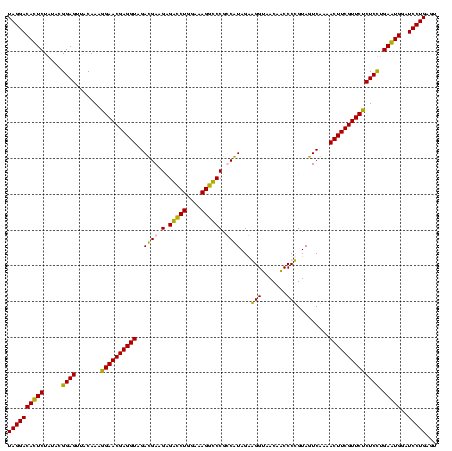
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -33.33 |
| Consensus MFE | -30.69 |
| Energy contribution | -28.50 |
| Covariance contribution | -2.19 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.754357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/320-440 ACUCAGGAUACAUUCAAGAGAGAACGAAGUUUCGACUACGGGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUA ....((((..(((((.((((.((((((.((...(((...(((((....)))))....((.((((((....))))).).))))).)).))))))))))(((......))).))))))))). ( -34.30) >Bcere.0 281279 120 + 5224283/320-440 ACUCAGGAUACAUUCAAGAGAGAACGAAGUUUCGACUACGGGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUA ....((((..(((((.((((.((((((.((...(((...(((((....)))))....((.((((((....))))).).))))).)).))))))))))(((......))).))))))))). ( -34.30) >Bsubt.0 636660 120 + 4214630/320-440 ACUCAGGAUCCACUCAGGAGAGAACGAAGUUUUGACUACAGGGCUGUUACCUCCUAUGGCGGGCCUUUCCAGACCUCUUCAUCUACCUCGUUCCUUUGUAACUCCGUACAGAGUGUCCUA ....((((..(((((.((((.((((((.((..(((....(((.((((((.......)))))).)))....((....)))))...)).))))))))))(((......))).))))))))). ( -31.90) >Blich.0 613689 120 + 4222334/320-440 ACUCAGGAUCCACUCAGGAGAGAACGAAGUUUUGACUACAGGGCUGUUACCUCCUAUGGCGGGCCUUUCCAGACCUCUUCAUCUACCUCGUUCUUUUGCAACUCCGUACAGAGUGUCCUA ....((((..(((((..((((((((((.((..(((....(((.((((((.......)))))).)))....((....)))))...)).))))))))))((......))...))))))))). ( -32.80) >consensus ACUCAGGAUACACUCAAGAGAGAACGAAGUUUCGACUACAGGGCUGUUACCCCCUACGACGGACCUUUCCAGACCGCUUCAUCUACCUCGUUCCUUUGUAACUCCGUACAGAAUGUCCUA ....((((..(((((.((((.((((((.((..(((.......(((((........))))).(((((....)))))...)))...)).))))))))))(((......))).))))))))). (-30.69 = -28.50 + -2.19)

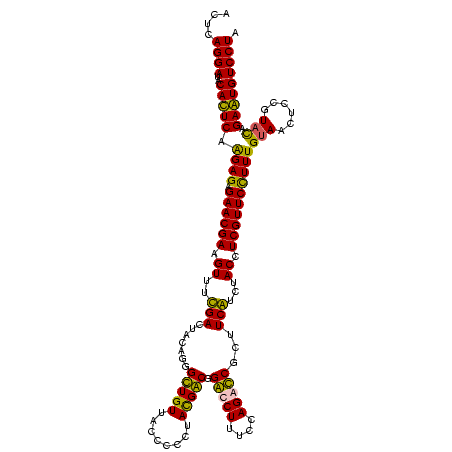
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.03 |
| Mean single sequence MFE | -41.45 |
| Consensus MFE | -41.41 |
| Energy contribution | -40.97 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.48 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/520-640 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAU ....((((((...((((..(.(....).).((((....))))...........)))).....)).))))(((((((..((((..(((((....)))))..))))......)))))))... ( -39.30) >Bcere.0 281279 120 + 5224283/520-640 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAU ....((((((...((((..(.(....).).((((....))))...........)))).....)).))))(((((((..((((..(((((....)))))..))))......)))))))... ( -41.90) >Bsubt.0 636660 120 + 4214630/520-640 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAU ....((((((...((((..(.(....).).((((....))))...........)))).....)).))))(((((((((((((..(((((....)))))..)))).)))...))))))... ( -42.30) >Blich.0 613689 120 + 4222334/520-640 ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAU ....((((((...((((..(.(....).).((((....))))...........)))).....)).))))(((((((((((((..(((((....)))))..)))).)))...))))))... ( -42.30) >consensus ACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAU ....((((((...((((..........((.((((....)))).))........)))).....)).))))(((((((..((((..(((((....)))))..))))......)))))))... (-41.41 = -40.97 + -0.44)

| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.40 |
| Mean single sequence MFE | -43.05 |
| Consensus MFE | -40.94 |
| Energy contribution | -39.75 |
| Covariance contribution | -1.19 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705076 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/600-720 CAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGUU ....((((.........((.((((.((((((((.(((....((((..((...((((....)))))).))))...))-)))))))))..))))))((....))...........))))... ( -40.50) >Bcere.0 281279 119 + 5224283/600-720 CAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGUU ....((((.....((((((.((((.((((((((.(((....((((..((...((((....)))))).))))...))-)))))))))..))))))((....))...))))....))))... ( -42.10) >Bsubt.0 636660 119 + 4214630/600-720 CAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGCA-GAAGAUGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCA ((..(((((....)))))..))..(((((((...((((......(((.((.......)))))(((..((((..(((-.....)))..)))))))((....))...))))...))))))). ( -44.60) >Blich.0 613689 120 + 4222334/600-720 CAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGUUUGAAAAGACGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCA ((..(((((....)))))..))..(((((((...((((......(((.((.......)))))(((..((((..((((....))))..)))))))((....))...))))...))))))). ( -45.00) >consensus CAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGCU_GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCA ((..(((((....)))))..))..(((((((...((((......(((.((.......)))))(((..((((..(((......)))..)))))))((....))...))))...))))))). (-40.94 = -39.75 + -1.19)


| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -46.60 |
| Consensus MFE | -45.90 |
| Energy contribution | -45.90 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/720-840 U-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGG .-..(((.((((((......)))..))).))).....(((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))) ( -45.90) >Bcere.0 281279 119 + 5224283/720-840 U-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGG .-..(((.((((((......)))..))).))).....(((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))) ( -45.90) >Bsubt.0 636660 120 + 4214630/720-840 UGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGG .((.(((.((((((......)))..))).))).))..(((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))) ( -47.30) >Blich.0 613689 120 + 4222334/720-840 UGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGG .((.(((.((((((......)))..))).))).))..(((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))) ( -47.30) >consensus U_AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGG ....(((.((((((......)))..))).))).....(((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))) (-45.90 = -45.90 + -0.00)

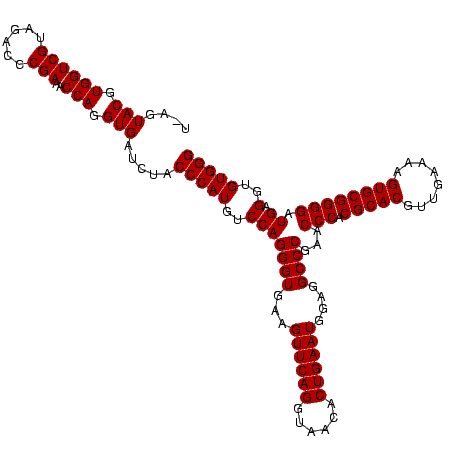
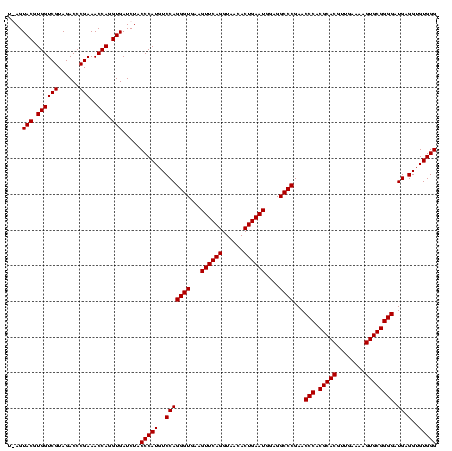
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.50 |
| Consensus MFE | -40.45 |
| Energy contribution | -39.95 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.864337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/760-880 AUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUC ((.((((((.(((.((((((......))))))((((.((((.((((((((.(((........)))........))))))))..))).)...))))..)))..)))))).))......... ( -42.10) >Bcere.0 281279 120 + 5224283/760-880 AUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUC ((.((((((.(((.((((((......))))))((((.((((.((((((((.(((........)))........))))))))..))).)...))))..)))..)))))).))......... ( -42.10) >Bsubt.0 636660 120 + 4214630/760-880 AUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUC ((.((((((.(((.((((((......))))))((..((((..((((((((.(((........)))........))))))))..))))......))..)))..)))))).))......... ( -40.90) >Blich.0 613689 120 + 4222334/760-880 AUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUC ((.((((((.(((.((((((......))))))((..((((..((((((((.(((........)))........))))))))..))))......))..)))..)))))).))......... ( -40.90) >consensus AUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUC ((.((((((.....((((((......)))))).((.(((((.((((((((((........)))))....)).))).)))))...(((.....)))..))...)))))).))......... (-40.45 = -39.95 + -0.50)

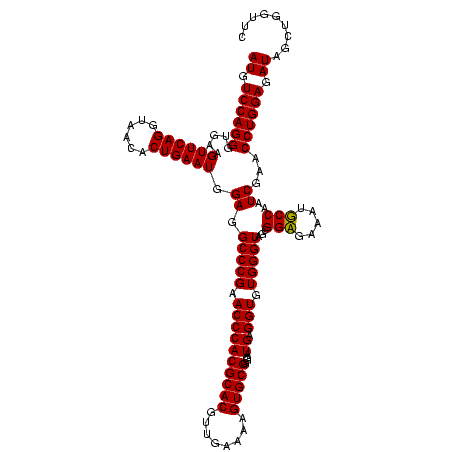
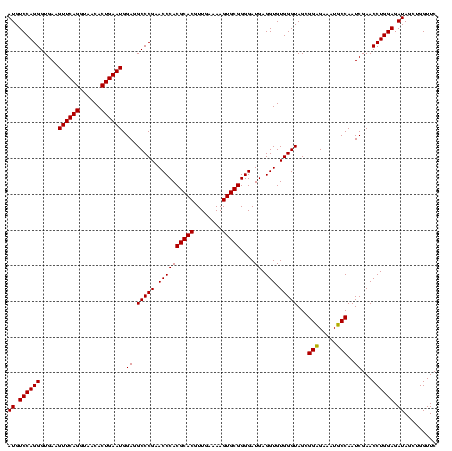
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -32.47 |
| Consensus MFE | -31.85 |
| Energy contribution | -31.85 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/760-880 GAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAU ...((((........((((((((..(((.....)))................((((((((........))))).))).))))))))...(((((......))))).......)))).... ( -33.90) >Bcere.0 281279 120 + 5224283/760-880 GAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAU ...((((........((((((((..(((.....)))................((((((((........))))).))).))))))))...(((((......))))).......)))).... ( -33.90) >Bsubt.0 636660 120 + 4214630/760-880 GAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAU ...((((........((((((((.(((..............)))........((((((((........))))).))).))))))))...(((((......))))).......)))).... ( -31.04) >Blich.0 613689 120 + 4222334/760-880 GAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAU ...((((........((((((((.(((..............)))........((((((((........))))).))).))))))))...(((((......))))).......)))).... ( -31.04) >consensus GAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAU ...((((........((((((((..((.......))................((((((((........))))).))).))))))))...(((((......))))).......)))).... (-31.85 = -31.85 + -0.00)

| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -39.30 |
| Consensus MFE | -39.40 |
| Energy contribution | -38.40 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.63 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764490 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/800-920 GAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAAGAGU ...(((.(((((........)))))))).((((((.((((....((((...))))....(((((.((......)))))))..))))...(((((....)))))))))))........... ( -38.40) >Bcere.0 281279 120 + 5224283/800-920 GAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAAGAGU ...(((.(((((........)))))))).((((((.((((....((((...))))....(((((.((......)))))))..))))...(((((....)))))))))))........... ( -38.40) >Bsubt.0 636660 119 + 4214630/800-920 GAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAG-GUAAGAGU ...(((.(((((........)))))))).((((((.((((((((..(((........)))..))))((((........))))))))...(((((....)))))))))))..-........ ( -40.20) >Blich.0 613689 119 + 4222334/800-920 GAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAG-GUAAGAGU ...(((.(((((........)))))))).((((((.((((((((..(((........)))..))))((((........))))))))...(((((....)))))))))))..-........ ( -40.20) >consensus GAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUCAAG_GUAAGAGU ...(((.(((((........)))))))).((((((.((((....(((.....)))....(((((.((......)))))))..))))...(((((....)))))))))))........... (-39.40 = -38.40 + -1.00)

| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -30.80 |
| Consensus MFE | -27.75 |
| Energy contribution | -28.75 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/800-920 ACUCUUACACUUAAGGCUAGCCCUAAAGCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUC ..............((.((((((.(((....))).))((((((((((..(((....)))....))))...)))))))))).)).........((((((((........))))).)))... ( -31.60) >Bcere.0 281279 120 + 5224283/800-920 ACUCUUACACUUAAGGCUAGCCCUAAAGCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUC ..............((.((((((.(((....))).))((((((((((..(((....)))....))))...)))))))))).)).........((((((((........))))).)))... ( -31.60) >Bsubt.0 636660 119 + 4214630/800-920 ACUCUUAC-CUUGAGGCUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUC ........-..(((((.((((......))))....(((((..........)))))(((...((........))...))).......))))).((((((((........))))).)))... ( -30.00) >Blich.0 613689 119 + 4222334/800-920 ACUCUUAC-CUUGAGGCUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUC ........-..(((((.((((......))))....(((((..........)))))(((...((........))...))).......))))).((((((((........))))).)))... ( -30.00) >consensus ACUCUUAC_CUUAAGGCUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUC ...........(((((.((((......))))....(((((((.((((..(((....)))....))))..)))))))..........))))).((((((((........))))).)))... (-27.75 = -28.75 + 1.00)

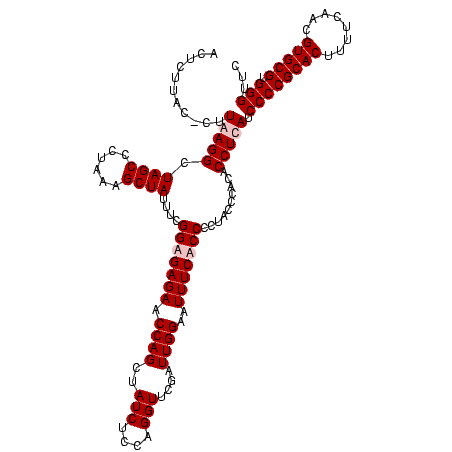
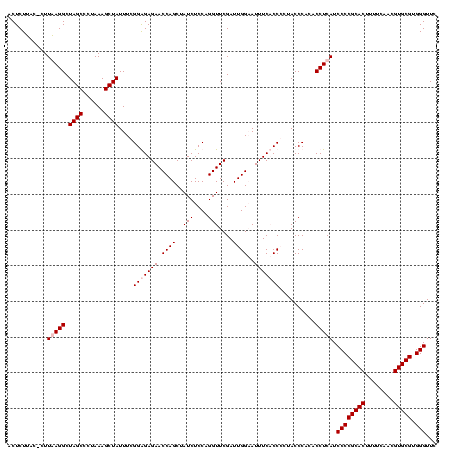
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -37.30 |
| Energy contribution | -36.80 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/1080-1200 GUGUAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGU .................((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..))))........ ( -37.10) >Bcere.0 281279 120 + 5224283/1080-1200 GUGUAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGU .................((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..))))........ ( -37.10) >Bsubt.0 636660 120 + 4214630/1080-1200 AUACGUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGU .(((.....))).....((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..))))........ ( -38.20) >Blich.0 613689 120 + 4222334/1080-1200 AUACGUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGU .(((.....))).....((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..))))........ ( -38.20) >consensus AUACAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGU .................((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..))))........ (-37.30 = -36.80 + -0.50)

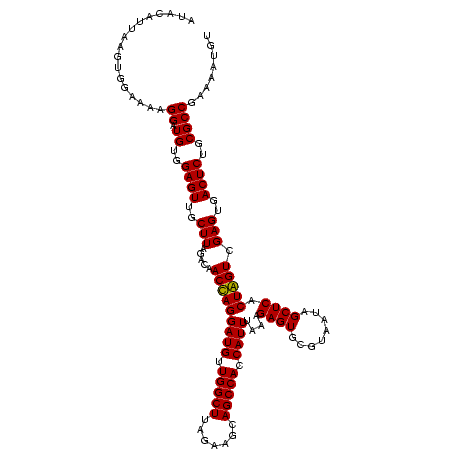
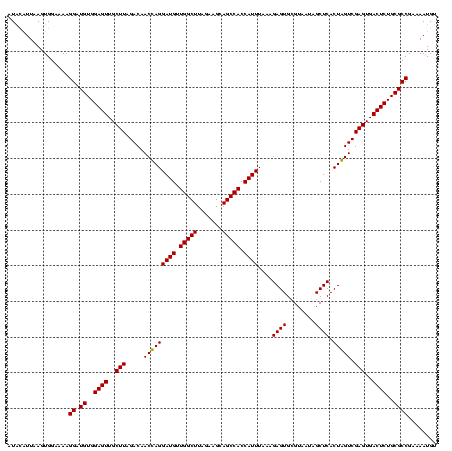
| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -40.62 |
| Consensus MFE | -40.37 |
| Energy contribution | -38.49 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.38 |
| SVM RNA-class probability | 0.993182 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/1200-1320 ACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACC-AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGA .(((.((((..............)))).)))(((((((((-...))))))))).........((.(((((((.((......(((((.((....)).)))))...)).))))))))).... ( -36.04) >Bcere.0 281279 120 + 5224283/1200-1320 ACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACCGAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGA .(((.((((..............)))).)))(((((((((....))))))))).........((.(((((((.((......(((((.((....)).)))))...)).))))))))).... ( -37.24) >Bsubt.0 636660 115 + 4214630/1200-1320 ACCGGGGCUAAACGUAUCACCGAAGCUGCGGACUGUUCU----U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGA .(((.((((...((......)).)))).)))...(((((----(-(..((....)).))))))).(((((((.(((.(...(((((.((....)).)))))).))).)))))))...... ( -44.50) >Blich.0 613689 116 + 4222334/1200-1320 ACCGGGGCUAAACGUAUCACCGAAGCUGCGGACUGUUCU----UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGA .(((.((((...((......)).)))).)))(((((((.----...))))))).........((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).... ( -44.70) >consensus ACCGGGGCUAAACACACCACCGAAGCUGCGGACUGAUAC____UGCGAACAGUGGUAGGAGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGA .(((.((((..............)))).)))((((((((......)))))))).........((.(((((((.((......(((((.((....)).)))))...)).))))))))).... (-40.37 = -38.49 + -1.88)

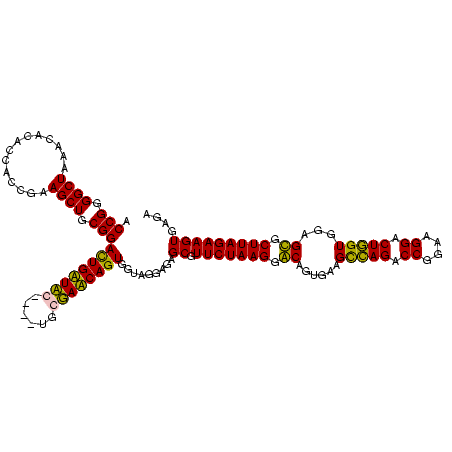
| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.68 |
| Mean single sequence MFE | -31.42 |
| Consensus MFE | -25.81 |
| Energy contribution | -27.12 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.82 |
| SVM decision value | 1.41 |
| SVM RNA-class probability | 0.951585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/1200-1320 UCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU-GGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGU ........(((((.((.(((((((.((....)).))(((......)))....(((.(((..........(((((((...-)))))))......))))))))))).)).)))))....... ( -27.79) >Bcere.0 281279 120 + 5224283/1200-1320 UCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUCGGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGU ........(((((.((.(((((((.((....)).))(((......)))....(((.(((..........(((((((....)))))))......))))))))))).)).)))))....... ( -29.39) >Bsubt.0 636660 115 + 4214630/1200-1320 UCUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCG-A----AGAACAGUCCGCAGCUUCGGUGAUACGUUUAGCCCCGGU ......(((((((.(((...((((.((....)).))))......))).)))))))............(((((((.-.----.)))))))(((..(((.((......))...)))..))). ( -31.90) >Blich.0 613689 116 + 4222334/1200-1320 UCUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCGCA----AGAACAGUCCGCAGCUUCGGUGAUACGUUUAGCCCCGGU ......(((((((.(((...((((.((....)).))))......))).)))))))............(((((((...----.)))))))(((..(((.((......))...)))..))). ( -36.60) >consensus UCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACAGCCCUUAGAACGCUCCCCUACCACUGAUACCA____AGAACAAUCCGCAGCUUCGGUGAUACAUUUAGCCCCGGU ........((((((((.(((((((.((....)).))(((......)))....(((.(((........(((((((((....)))))))))....))))))))))).))))))))....... (-25.81 = -27.12 + 1.31)

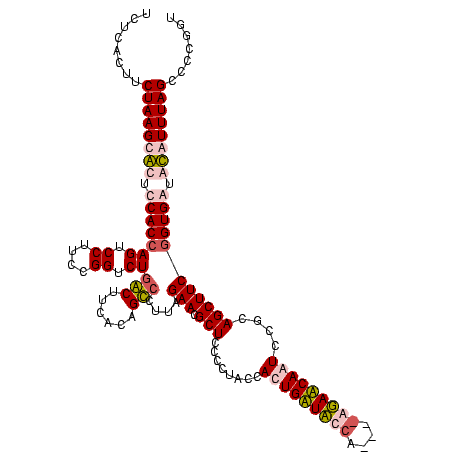
| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -39.12 |
| Energy contribution | -37.50 |
| Covariance contribution | -1.62 |
| Combinations/Pair | 1.13 |
| Mean z-score | -3.21 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.20 |
| SVM RNA-class probability | 0.998733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/1240-1360 -AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCGUC -..((.(((((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))...))).)).))...((((((.......)))))) ( -35.70) >Bcere.0 281279 120 + 5224283/1240-1360 GAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCGUC ...((.(((((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))...))).)).))...((((((.......)))))) ( -35.70) >Bsubt.0 636660 116 + 4214630/1240-1360 ---U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGGGGUGAGAAUCCCCUC ---(-((.((...((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))).)).))).........((((((.......)))))) ( -44.30) >Blich.0 613689 117 + 4222334/1240-1360 ---UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGGGGUGAGAAUCCCCUC ---.((...((.(((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))))...))....))....((((((.......)))))) ( -44.70) >consensus ___UGCGAACAGUGGUAGGAGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCCUC ..........(.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).).............((((((.......)))))) (-39.12 = -37.50 + -1.62)


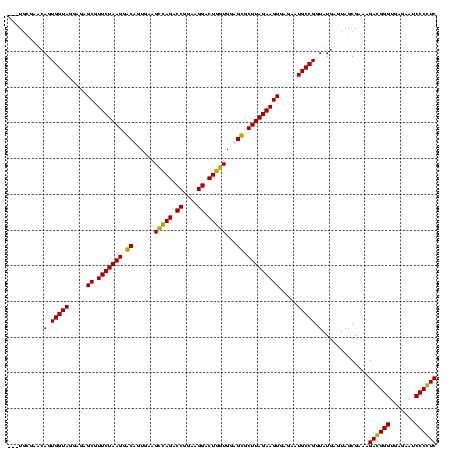
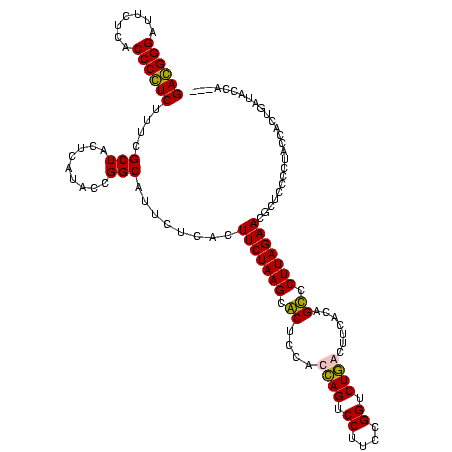
| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.61 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -25.77 |
| Energy contribution | -24.90 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.995911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/1240-1360 GACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU- ((((((.......))))))....(((.........)))........(((((((.((.....(((.((....)).)))........)).)))))))........................- ( -22.72) >Bcere.0 281279 120 + 5224283/1240-1360 GACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUC ((((((.......))))))....(((.........)))........(((((((.((.....(((.((....)).)))........)).)))))))......................... ( -22.72) >Bsubt.0 636660 116 + 4214630/1240-1360 GAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCG-A--- ((((((.......))))))....(((.........)))........(((((((.(((...((((.((....)).))))......))).)))))))....................-.--- ( -29.70) >Blich.0 613689 117 + 4222334/1240-1360 GAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCGCA--- ((((((.......))))))....(((.........)))........(((((((.(((...((((.((....)).))))......))).)))))))......................--- ( -29.10) >consensus GACGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACAGCCCUUAGAACGCUCCCCUACCACUGAUACCA___ ((((((.......))))))....(((.........)))........(((((((.((....((((.((....)).)))).......)).)))))))......................... (-25.77 = -24.90 + -0.87)

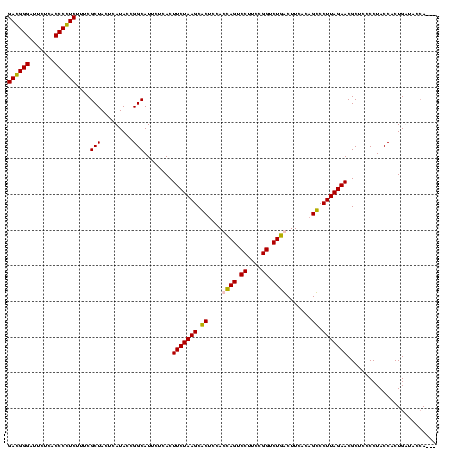
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -41.00 |
| Energy contribution | -40.50 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.55 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/1320-1440 CUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAU ....(((((((((((((.(((((((...........))))))).)).).)))((((...))))......))))..(((.(((((((.......))))))).)))...........))).. ( -41.50) >Bcere.0 281279 120 + 5224283/1320-1440 CUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAU ....(((((((((((((.(((((((...........))))))).)).).)))((((...))))......))))..(((.(((((((.......))))))).)))...........))).. ( -41.50) >Bsubt.0 636660 120 + 4214630/1320-1440 CUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAU ....(((....((((((.(((((((...........))))))).)).).)))..(((.(((...))).)))....(((.(((((((.......))))))).)))...........))).. ( -40.90) >Blich.0 613689 120 + 4222334/1320-1440 CUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAU ....(((....((((((.(((((((...........))))))).)).).)))..(((.(((...))).)))....(((.(((((((.......))))))).)))...........))).. ( -40.90) >consensus CUACGCCUGUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAU ....(((....((((((.(((((((...........))))))).)).).)))..(((.(((...))).)))....(((.(((((((.......))))))).)))...........))).. (-41.00 = -40.50 + -0.50)

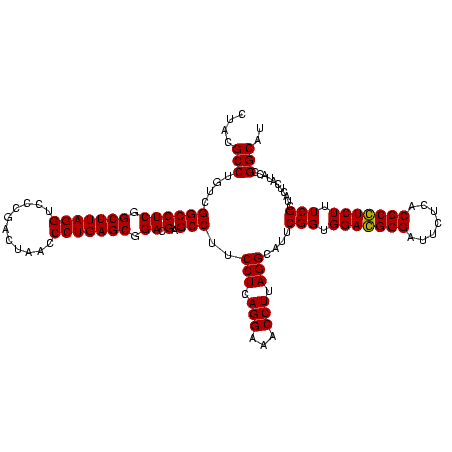
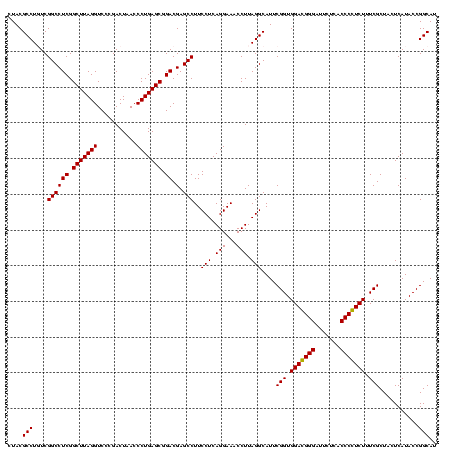
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -32.18 |
| Consensus MFE | -29.91 |
| Energy contribution | -28.66 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/1440-1560 UUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGC ((((((((......))).((((((((..........))))((...............))))))...............)))))...(((((.(((((........)))))...))))).. ( -28.86) >Bcere.0 281279 120 + 5224283/1440-1560 UUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGC ((((((((......))).((((((((..........))))((...............))))))...............)))))...(((((.(((((........)))))...))))).. ( -28.86) >Bsubt.0 636660 120 + 4214630/1440-1560 UUCCCAGCCUAACUGCUUGGACGCGGAUAUCCAAUACCGCGCUUACCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAAUGGUGAGGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGC .(((.(((......))).)))(((((..........)))))................((.(((.((((((.....))))).).)))(((((.(((((........)))))...))))))) ( -35.50) >Blich.0 613689 120 + 4222334/1440-1560 UUCCCAGCCUAACUGCUUGGACGCGGAUAUCCAAUACCGCGCUUGCCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAAUGGUGAGGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGC .(((.(((......))).)))(((((..........)))))................((.(((.((((((.....))))).).)))(((((.(((((........)))))...))))))) ( -35.50) >consensus UUACCAGCCUAACUGCUUGGACGCGCACAACCAAUACCACGCUUCCCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGC ((((((((......))).((((((((..........))))((...............))))))...............)))))...(((((.(((((........)))))...))))).. (-29.91 = -28.66 + -1.25)

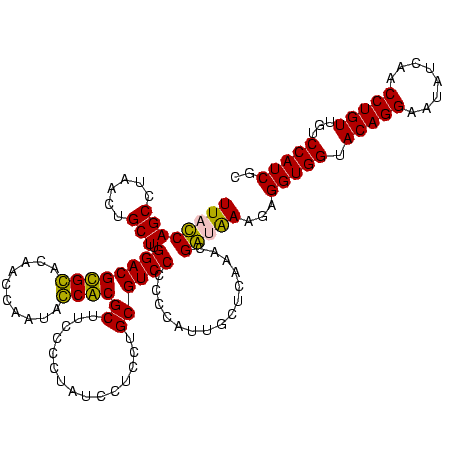
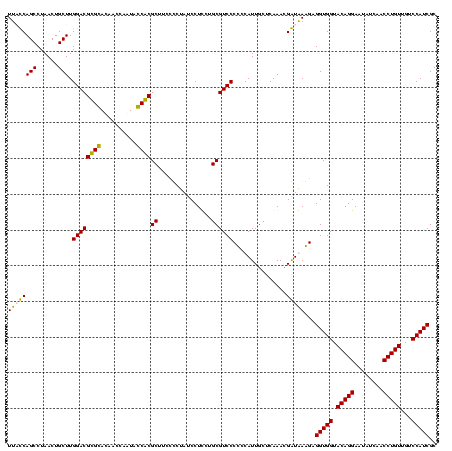
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 87.48 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -25.76 |
| Energy contribution | -23.76 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.06 |
| Structure conservation index | 1.01 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/1480-1600 CCAUCACAGCUCAGCCUUCACGAUAAGCGGAUUUGCCUACUUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGA .......(((...........((((((..(......)..))))))(((......))).((((((((..........))))((...............)))))).......)))....... ( -21.16) >Bcere.0 281279 120 + 5224283/1480-1600 CCAUCACAGCUCAGCCUUUACGAUAAGCGGAUUUGCCUACUUAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGA .......(((...........((((((..(......)..))))))(((......))).((((((((..........))))((...............)))))).......)))....... ( -21.16) >Bsubt.0 636660 119 + 4214630/1480-1600 CCAUCACAGCUCAGCCUU-AUGGGAAACGGAUUUGCCUAUUUCCCAGCCUAACUGCUUGGACGCGGAUAUCCAAUACCGCGCUUACCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAAUGG ((((...(((.(((....-.(((((((..(......)..)))))))......)))...((((((((..........))))((...............)))))).......)))...)))) ( -29.36) >Blich.0 613689 119 + 4222334/1480-1600 CCAUCACAGCUCAGCCUU-AUGGGAAACGGAUUUGCCUAUUUCCCAGCCUAACUGCUUGGACGCGGAUAUCCAAUACCGCGCUUGCCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAAUGG ((((...(((.(((....-.(((((((..(......)..)))))))......)))...(((((((((((..(((........)))...)))))....)))))).......)))...)))) ( -30.80) >consensus CCAUCACAGCUCAGCCUU_ACGAGAAACGGAUUUGCCUACUUACCAGCCUAACUGCUUGGACGCGCACAACCAAUACCACGCUUCCCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAACGA .......(((.(((......(((((((..(......)..)))))))......)))...((((((((..........))))((...............)))))).......)))....... (-25.76 = -23.76 + -2.00)

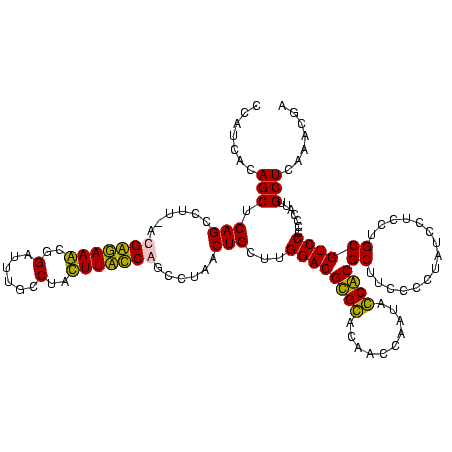
| Location | 247,889 – 247,998 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -29.33 |
| Energy contribution | -32.95 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.645504 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 109 + 5227293/1760-1880 GUAACUUCGGGAGAAGGGGUGCUUUCUUAACG-----------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAG ...(((((.(((((..(((((((((((....)-----------))))))............))))..((...(((((.....)))))..))))))).....(((....)))))))).... ( -33.50) >Bcere.0 281279 109 + 5224283/1760-1880 GUAACUUCGGGAGAAGGGGUGCUUUCUUAACG-----------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAG ...(((((.(((((..(((((((((((....)-----------))))))............))))..((...(((((.....)))))..))))))).....(((....)))))))).... ( -33.50) >Bsubt.0 636660 120 + 4214630/1760-1880 GUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAG ...(((((.(((((..(.(.(((((.((.((((....)))))).)))))).).(((.....((.(((....)))....)).....)))...))))).....(((....)))))))).... ( -42.10) >Blich.0 613689 120 + 4222334/1760-1880 GUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAG ...(((((.(((((..(.(.(((((.((.((((....)))))).)))))).).(((.....((.(((....)))....)).....)))...))))).....(((....)))))))).... ( -42.10) >consensus GUAACUUCGGGAGAAGGGGUGCUCUCUUAACG___________GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAG ...(((((.(((((...(((((((((...((((....))))..)))))).((..(((((((((....))..)))))))))....)).)...))))).....(((....)))))))).... (-29.33 = -32.95 + 3.62)

| Location | 247,889 – 247,998 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -27.25 |
| Energy contribution | -28.75 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 109 + 5227293/1760-1880 CUAUACUUCGCCUUACGGCUUCGCAGAGAGCUGUGUUUUUGCUAAACAGUCGCUUGGGCCUAUUCACUGCGGCUUUC-----------CGUUAAGAAAGCACCCCUUCUCCCGAAGUUAC ....((((((((....)))..(((((.(((..(.((((..((.........))..)))))..))).)))))((((((-----------......))))))............)))))... ( -29.60) >Bcere.0 281279 109 + 5224283/1760-1880 CUAUACUUCGCCUUACGGCUUCGCAGAGAGCUGUGUUUUUGCUAAACAGUCGCUUGGGCCUAUUCACUGCGGCUUUC-----------CGUUAAGAAAGCACCCCUUCUCCCGAAGUUAC ....((((((((....)))..(((((.(((..(.((((..((.........))..)))))..))).)))))((((((-----------......))))))............)))))... ( -29.60) >Bsubt.0 636660 120 + 4214630/1760-1880 CUAUACUUCGCCUUACGGCUUCGCAGAGACCUGUGUUUUUGCUAAACAGUCGCCUGGGCCUAUUCACUGCGGCUCUCUCGGGCUUGCACCCUAACAGAGCACCCCUUCUCCCGAAGUUAC ....(((((((((..(((((..((((((((....)))))))).....)))))...))))............(((((...(((......)))....)))))............)))))... ( -33.80) >Blich.0 613689 120 + 4222334/1760-1880 CUAUACUUCGCCUUACGGCUUCGCAGAGACCUGUGUUUUUGCUAAACAGUCGCCUGGGCCUAUUCACUGCGGCUCUCUCGGGCUUGCACCCUAACAGAGCACCCCUUCUCCCGAAGUUAC ....(((((((((..(((((..((((((((....)))))))).....)))))...))))............(((((...(((......)))....)))))............)))))... ( -33.80) >consensus CUAUACUUCGCCUUACGGCUUCGCAGAGACCUGUGUUUUUGCUAAACAGUCGCCUGGGCCUAUUCACUGCGGCUCUC___________CCCUAACAAAGCACCCCUUCUCCCGAAGUUAC ....((((((((....)))..(((((.((.((((...........))))))((....)).......)))))((((((..(((......)))...))))))............)))))... (-27.25 = -28.75 + 1.50)

| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -39.55 |
| Energy contribution | -41.05 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/1840-1960 AAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAAC .....(((..((((.(((...(((....)))...))).)))).))).((((.....))))((((..((((..((...((((((.((....))..))))))...))..))))))))..... ( -42.10) >Bcere.0 281279 120 + 5224283/1840-1960 AAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAAC .....(((..((((.(((...(((....)))...))).)))).))).((((.....))))((((..((((..((...((((((.((....))..))))))...))..))))))))..... ( -42.10) >Bsubt.0 636660 120 + 4214630/1840-1960 AAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAAC ............((.(((...(((....)))...))).))((((........))))..((((((..((((..((...((((((.((....))..))))))...))..))))))))))... ( -43.30) >Blich.0 613689 120 + 4222334/1840-1960 AAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAAC ............((.(((...(((....)))...))).))((((........))))..((((((..((((..((...((((((.((....))..))))))...))..))))))))))... ( -43.30) >consensus AAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAAGAGCGCUUAGCGUAAGCGAAGCUCCGAACUGAAGCCCCAGUAAAC .........(((((((((...(((....)))...))..)))))))..((((.....))))((((..((((..((...((((((.((....))..))))))...))..))))))))..... (-39.55 = -41.05 + 1.50)

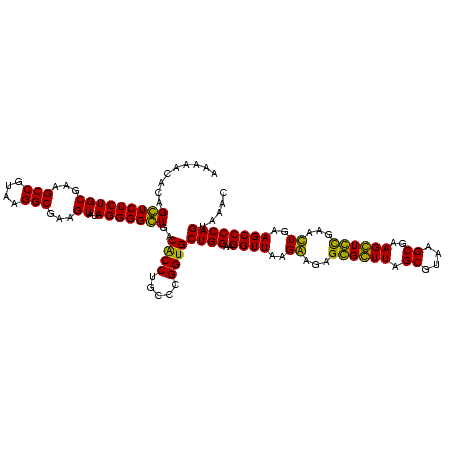
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -43.95 |
| Consensus MFE | -42.55 |
| Energy contribution | -42.55 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.857710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/1880-2000 GGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC (((((...(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...))))......))))) ( -42.80) >Bcere.0 281279 120 + 5224283/1880-2000 GGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC (((((...(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...))))......))))) ( -42.80) >Bsubt.0 636660 120 + 4214630/1880-2000 GGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC ..(((...(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...)))))))........ ( -45.10) >Blich.0 613689 120 + 4222334/1880-2000 GGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC ..(((...(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...)))))))........ ( -45.10) >consensus GGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAAGAGCGCUUAGCGUAAGCGAAGCUCCGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCC (((.((..(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...))))..))....))) (-42.55 = -42.55 + 0.00)

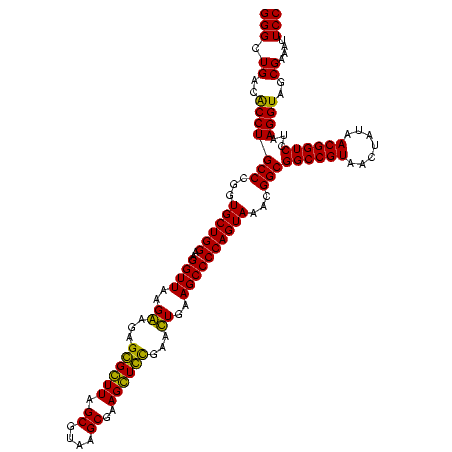
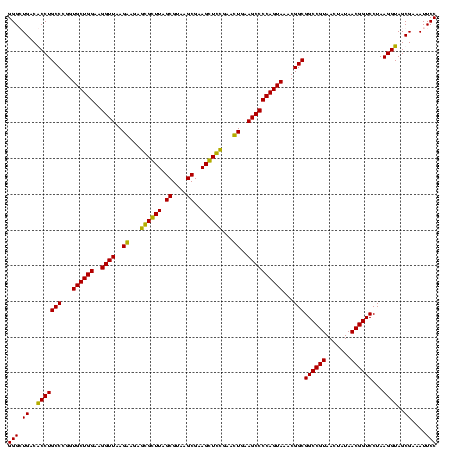
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -36.95 |
| Consensus MFE | -36.25 |
| Energy contribution | -36.25 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.659563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2000-2120 UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUGGUUGAGACAGUGCCCAAAUCGUUACACCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.(((((((((((((........)))))................(((((.....)))))..))))).....(.....).....))))))).)))))............ ( -36.10) >Bcere.0 281279 120 + 5224283/2000-2120 UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUGGUUGAGACAGUGCCCAAAUCGUUACACCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.(((((((((((((........)))))................(((((.....)))))..))))).....(.....).....))))))).)))))............ ( -36.10) >Bsubt.0 636660 120 + 4214630/2000-2120 UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAGAUCGUUGCGCCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.((((((.....((((...((...((((((....(((((.((((...)))).))))).))))))..))....))))...)))))))))).)))))............ ( -37.80) >Blich.0 613689 120 + 4222334/2000-2120 UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAGAUCGUUGCGCCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.((((((.....((((...((...((((((....(((((.((((...)))).))))).))))))..))....))))...)))))))))).)))))............ ( -37.80) >consensus UCUUUCCGUCCUGUCGCGGGUAACCUGCAUCUUCACAGGUACUAUAAUUUCACCGAGUCUCUCGUUGAGACAGUGCCCAAAUCGUUACACCUUUCGUGCGGGUCGGAACUUACCCGACAA ...(((((.((((.(((((((((((((........)))))................(((((.....)))))..))))).....(.....).....))))))).)))))............ (-36.25 = -36.25 + 0.00)

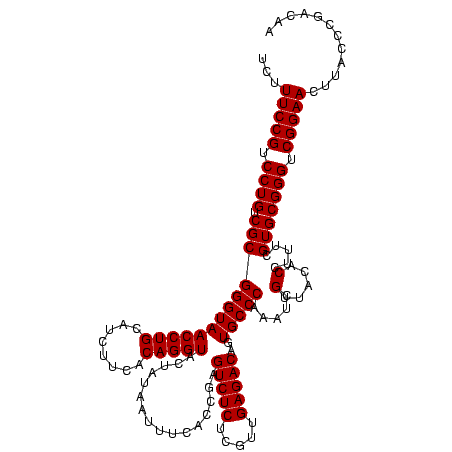
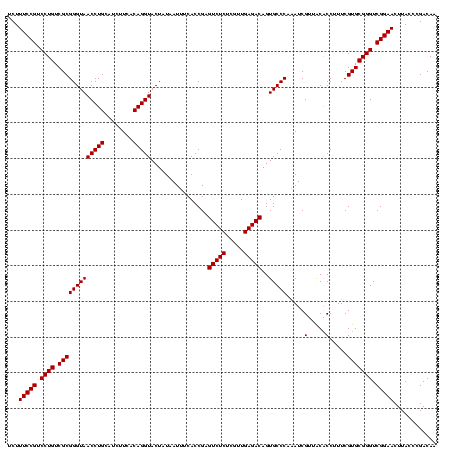
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -49.83 |
| Consensus MFE | -51.00 |
| Energy contribution | -48.12 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.04 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.26 |
| SVM RNA-class probability | 0.991355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2160-2280 ACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGG ....((.((((((..(((((((..(((((....((((((((....)))))))).))))).)))((((....)))))))).)))))).))............((((((.....)))))).. ( -49.40) >Bcere.0 281279 120 + 5224283/2160-2280 ACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGG ....((.((((((..(((((((..(((((....((((((((....)))))))).))))).)))((((....)))))))).)))))).))............((((((.....)))))).. ( -49.40) >Bsubt.0 636660 120 + 4214630/2160-2280 ACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGG ..........(((.((((.((((((((((....((((((((....)))))))).))))).))(((((....)))))))).)))).)))..........((.((((((.....)))))))) ( -51.00) >Blich.0 613689 120 + 4222334/2160-2280 ACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGG ..........(((.((((.((((((((((....((((((((....)))))))).))))).))(((((....)))))))).)))).)))..........((.((((((.....)))))))) ( -49.50) >consensus ACAGCUUGUACAGGAUAGGCAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCACCGGUGGGAUACCACCCUGACUGUAUUGAAAUUCUAACCCACGGCCCUUAUCGACCCGGG .......((((((..(((...((.(((((....((((((((....)))))))).))))).))(((((....)))))))).))))))............((.((((((.....)))))))) (-51.00 = -48.12 + -2.88)

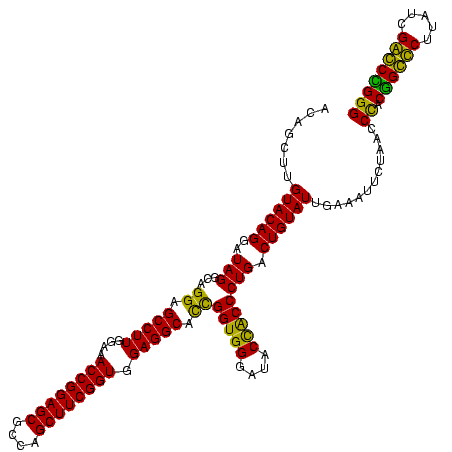
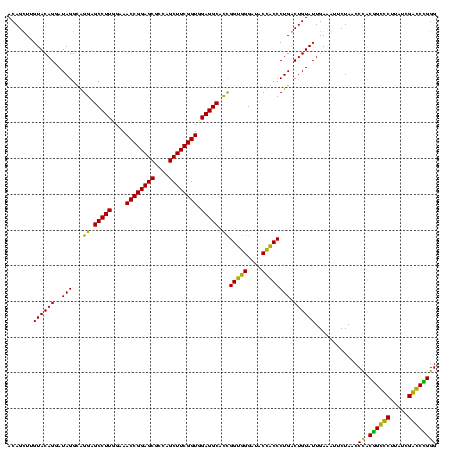
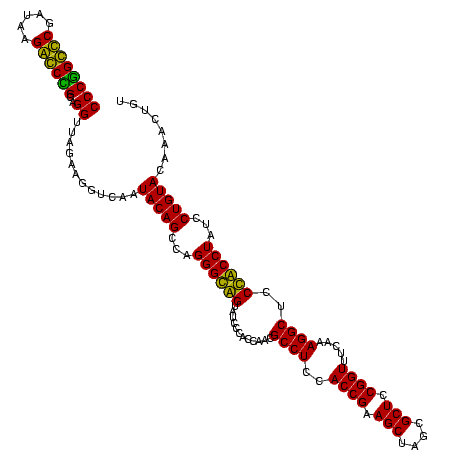
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -41.23 |
| Consensus MFE | -38.94 |
| Energy contribution | -36.31 |
| Covariance contribution | -2.62 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2160-2280 CCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGU ((((((((.....))))))..))............(((((...(((((((.....)))...((((..((((.(((....))).)))).....))))....))))...)))))........ ( -38.30) >Bcere.0 281279 120 + 5224283/2160-2280 CCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGU ((((((((.....))))))..))............(((((...(((((((.....)))...((((..((((.(((....))).)))).....))))....))))...)))))........ ( -38.30) >Bsubt.0 636660 120 + 4214630/2160-2280 CCCCGCCCGAUAAGGGCGGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCUGUACAAGCUGU ((((((((.....)))))).))..............(((((((((((((.(((.....)))((((.....((((((((....))))))))..))))......)))))))).....))))) ( -44.90) >Blich.0 613689 120 + 4222334/2160-2280 CCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCUGUACAAGCUGU ((((((((.....)))))).))..............(((((((((((((.(((.....)))((((.....((((((((....))))))))..))))......)))))))).....))))) ( -43.40) >consensus CCCGGCCCGAUAAGACCCGCAGGUUAGAAGGUCAAUACAGCCAGGGCAGUAUCCCACCAACGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCACCUAUCCUGUACAAACUGU ((((((((.....))))))..))............(((((...((((((............((((..((((.(((....))).)))).....))))..))))))...)))))........ (-38.94 = -36.31 + -2.62)

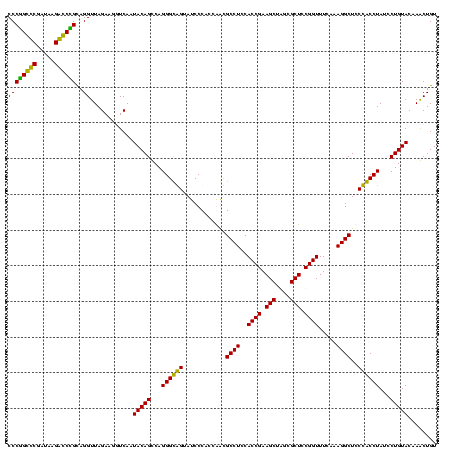
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -46.73 |
| Consensus MFE | -45.25 |
| Energy contribution | -44.62 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2240-2360 CUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAU ..........(((((.......(((((.....)))))(((((((...)))...(((((.....(((((...)))))((((((...........))))))))))).....))))..))))) ( -43.80) >Bcere.0 281279 120 + 5224283/2240-2360 CUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAU ..........(((((.......(((((.....)))))(((((((...)))...(((((.....(((((...)))))((((((...........))))))))))).....))))..))))) ( -43.80) >Bsubt.0 636660 120 + 4214630/2240-2360 CUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAU (((....((((((...((((.((((((.....)))))).).(((...))).)))((((.....(((((...)))))((((((...........)))))))))).))))))....)))... ( -50.40) >Blich.0 613689 120 + 4222334/2240-2360 CUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAU (((....((((((...((((.((((((.....)))))).).(((...))).)))((((.....(((((...)))))((((((...........)))))))))).))))))....)))... ( -48.90) >consensus CUGUAUUGAAAUUCUAACCCACGGCCCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAU (((....((((((...(((..((((((.....))))))(....).......)))((((.....(((((...)))))((((((...........)))))))))).))))))....)))... (-45.25 = -44.62 + -0.62)

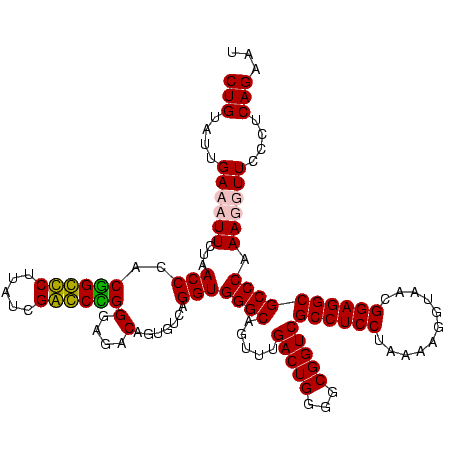
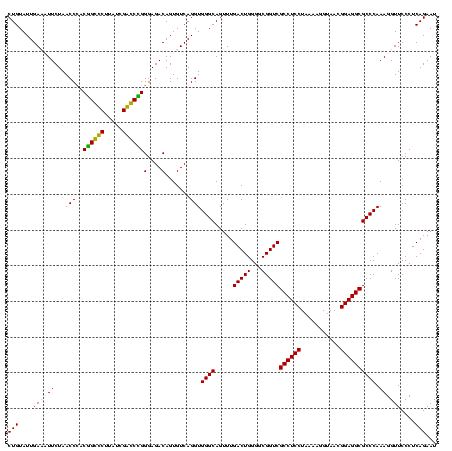
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.06 |
| Mean single sequence MFE | -45.73 |
| Consensus MFE | -45.20 |
| Energy contribution | -43.58 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.65 |
| SVM RNA-class probability | 0.996062 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2240-2360 AUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAG (((((((((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....))))).....))))))).......... ( -44.10) >Bcere.0 281279 120 + 5224283/2240-2360 AUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAG (((((((((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....))))).....))))))).......... ( -44.10) >Bsubt.0 636660 120 + 4214630/2240-2360 AUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCCGCCCGAUAAGGGCGGCGGGUUAGAAGGUCAAUACAG ...((((..((.(((((((((((((((...........))))))(((.......))).....))))).(((((.(((.....((((((.....))))))))))))))))))))..).))) ( -48.10) >Blich.0 613689 120 + 4222334/2240-2360 AUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAG ...((((..((.(((((((((((((((...........))))))(((.......))).....))))).(((((.(((.....((((((.....))))))))))))))))))))..).))) ( -46.60) >consensus AUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGCCCGAUAAGACCCGCAGGUUAGAAGGUCAAUACAG .((((((((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....))))).....))))))........... (-45.20 = -43.58 + -1.63)

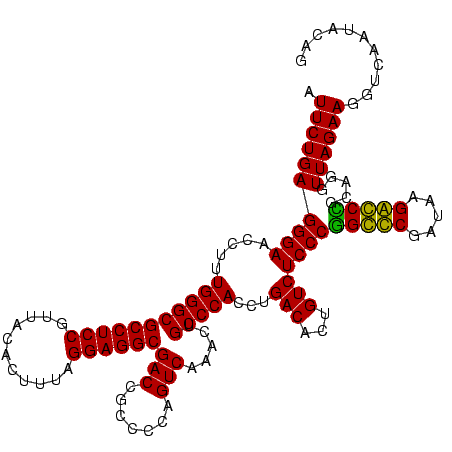
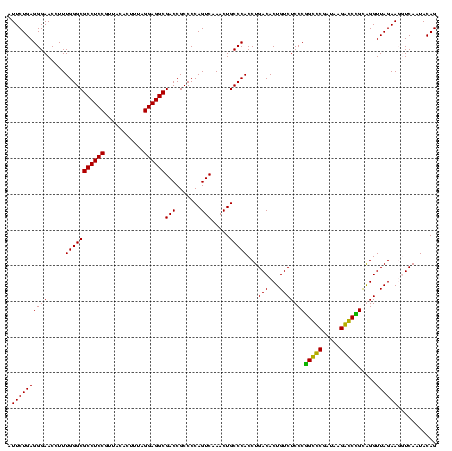
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -45.15 |
| Consensus MFE | -45.65 |
| Energy contribution | -45.15 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.51 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.83 |
| SVM RNA-class probability | 0.979305 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2280-2400 AGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGC .(((...)))...(((((.....(((((...)))))((((((...........)))))))))))...(((((((..(((((((.....))))))).....((((....)))).))))))) ( -44.90) >Bcere.0 281279 120 + 5224283/2280-2400 AGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGC .(((...)))...(((((.....(((((...)))))((((((...........)))))))))))...(((((((..(((((((.....))))))).....((((....)))).))))))) ( -44.90) >Bsubt.0 636660 120 + 4214630/2280-2400 AGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGC .(((...)))...(((((.....(((((...)))))((((((...........)))))))))))...(((((((..(((((((.....))))))).....((((....)))).))))))) ( -45.40) >Blich.0 613689 120 + 4222334/2280-2400 AGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGC .(((...)))...(((((.....(((((...)))))((((((...........)))))))))))...(((((((..(((((((.....))))))).....((((....)))).))))))) ( -45.40) >consensus AGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGCAAAGGCACAAGGGAGC .(((...)))...(((((.....(((((...)))))((((((...........)))))))))))...(((((((..(((((((.....))))))).....((((....)))).))))))) (-45.65 = -45.15 + -0.50)

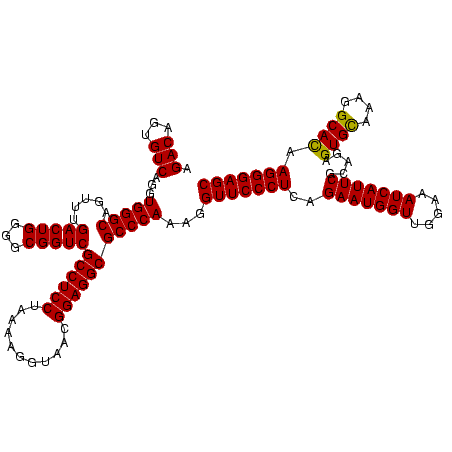
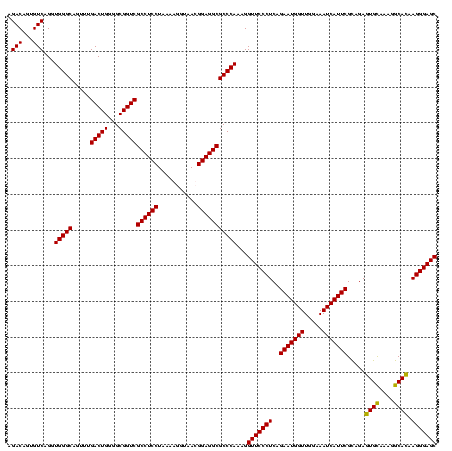
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -34.40 |
| Energy contribution | -34.90 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956300 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2280-2400 GCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCU ..((((((((((....)))......(((((.........)))))))))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))). ( -36.80) >Bcere.0 281279 120 + 5224283/2280-2400 GCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCU ..((((((((((....)))......(((((.........)))))))))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))). ( -36.80) >Bsubt.0 636660 120 + 4214630/2280-2400 GCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCU ..(((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))). ( -36.20) >Blich.0 613689 120 + 4222334/2280-2400 GCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCU ..(((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))). ( -36.20) >consensus GCUCCCUUAUGCCUUUACACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCU ..(((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))). (-34.40 = -34.90 + 0.50)

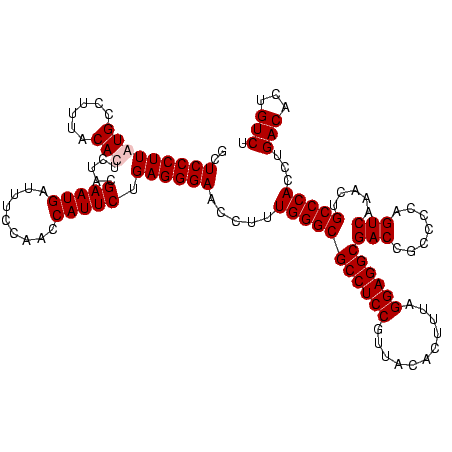
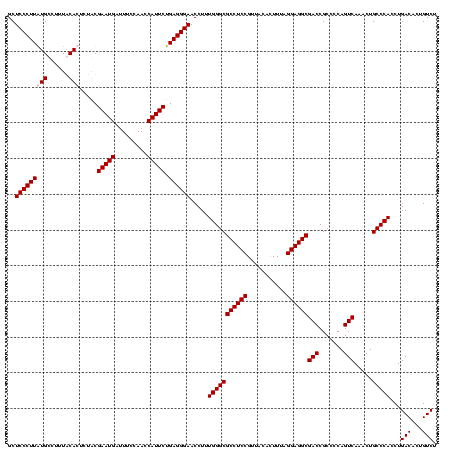
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -40.10 |
| Consensus MFE | -40.25 |
| Energy contribution | -39.75 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.27 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.960824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2320-2440 CCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCG ......((((.......))))....(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))). ( -40.20) >Bcere.0 281279 120 + 5224283/2320-2440 CCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCG ......((((.......))))....(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))). ( -40.20) >Bsubt.0 636660 120 + 4214630/2320-2440 CCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCG .............((......))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))). ( -40.00) >Blich.0 613689 120 + 4222334/2320-2440 CCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCG .............((......))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))). ( -40.00) >consensus CCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCG .............((......))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))). (-40.25 = -39.75 + -0.50)

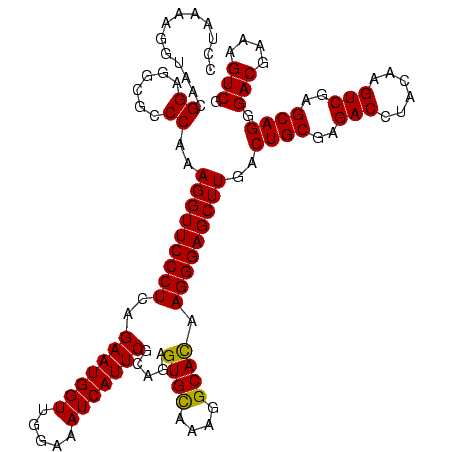
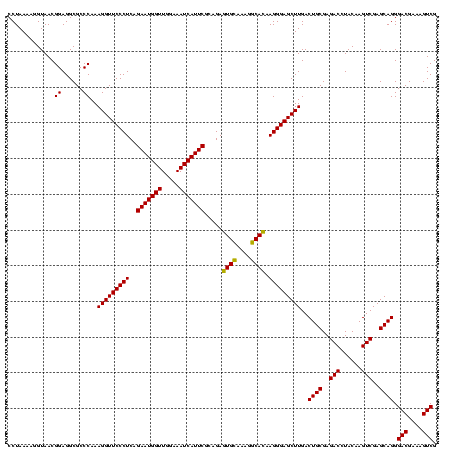
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -45.70 |
| Consensus MFE | -45.70 |
| Energy contribution | -45.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.89 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.587820 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2480-2600 UCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... ( -45.70) >Bcere.0 281279 120 + 5224283/2480-2600 UCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... ( -45.70) >Bsubt.0 636660 120 + 4214630/2480-2600 UCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... ( -45.70) >Blich.0 613689 120 + 4222334/2480-2600 UCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... ( -45.70) >consensus UCAACGGAUAAAAGCUACCCCGGGGAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAA .....((((....((((((((.(((((.....(((.((((((((...((.......))...))))))))..)))....(((((......))))).))))).)))..)))))..))))... (-45.70 = -45.70 + -0.00)

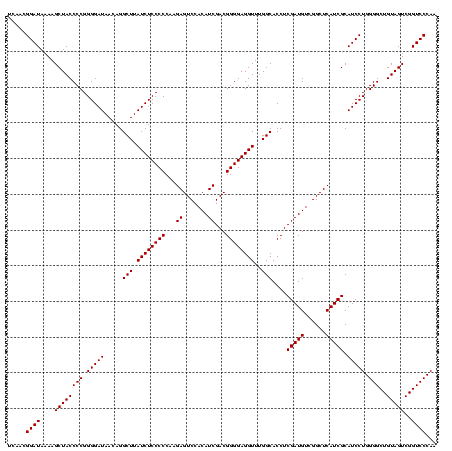
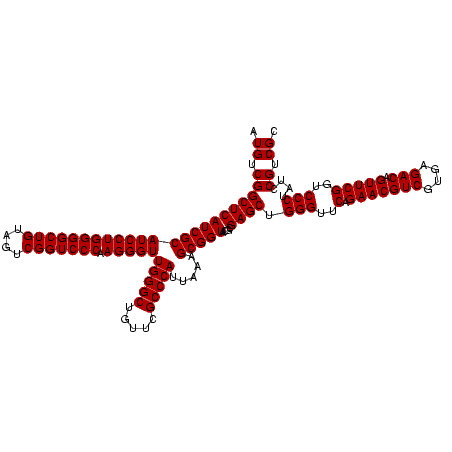
| Location | 247,889 – 248,009 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -53.00 |
| Consensus MFE | -49.75 |
| Energy contribution | -52.75 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.729586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 120 + 5227293/2560-2680 AUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGU (((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).))) ( -53.30) >Bcere.0 281279 120 + 5224283/2560-2680 AUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGU (((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).))) ( -53.30) >Bsubt.0 636660 120 + 4214630/2560-2680 AUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGC ..(.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).).. ( -52.70) >Blich.0 613689 120 + 4222334/2560-2680 AUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGC ..(.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).).. ( -52.70) >consensus AUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCGC .((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))).(((..(.(((((((....))).)))))..)))....)).)). (-49.75 = -52.75 + 3.00)

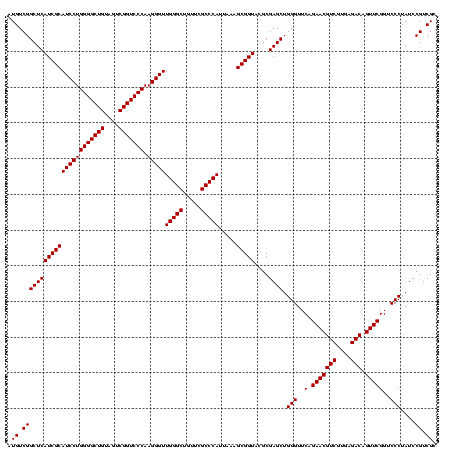
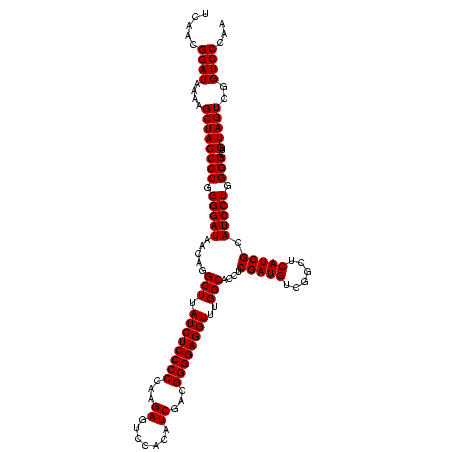
| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -31.48 |
| Energy contribution | -31.22 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/2800-2920 UGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUAGC-GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGU .(((..(.((......)).)..))).((((((.......(((((..(.((((-....)))))..)))))(((((.......)))))...........)))).))..(((((....))))) ( -35.70) >Bcere.0 281279 119 + 5224283/2800-2920 UGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUAGC-GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGU .(((..(.((......)).)..))).((((((.......(((((..(.((((-....)))))..)))))(((((.......)))))...........)))).))..(((((....))))) ( -35.70) >Bsubt.0 636660 119 + 4214630/2800-2920 UGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUUCC-GCAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAU .(((..(.((......)).)..))).(((((((.(((..(((((..(.((((-....)))))..)))))(((((.......))))).......)))))))).))..(((((....))))) ( -37.80) >Blich.0 613689 120 + 4222334/2800-2920 UGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUUCCUAUAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAU .(((..(.((......)).)..))).(((((((.(((..(((((..(.((((.....)))))..)))))(((((.......))))).......)))))))).))..(((((....))))) ( -34.30) >consensus UGCUGAAAGCAUCUAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUACC_GUAAGCAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCCGAGGUGGAAGCAUGGCGACACAU .(((..(.((......)).)..))).(((((((.(((..(((((..(.((((.....)))))..)))))(((((.......))))).......)))))))).))..(((((....))))) (-31.48 = -31.22 + -0.25)

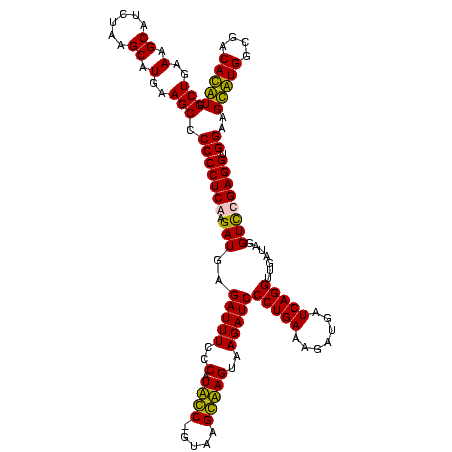
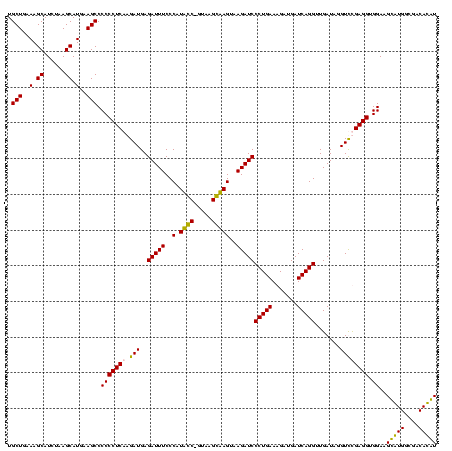
| Location | 247,889 – 248,008 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -33.35 |
| Energy contribution | -30.85 |
| Covariance contribution | -2.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.46 |
| Structure conservation index | 1.07 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 119 + 5227293/2800-2920 ACAUGUCACCAUGCUUCCACCUCGAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCA .((((....))))..(((.((((((.........(((((.......)))))..(((((.((((....-))))))))).........)))))))))(((....((......))....))). ( -31.00) >Bcere.0 281279 119 + 5224283/2800-2920 ACAUGUCACCAUGCUUCCACCUCGAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCA .((((....))))..(((.((((((.........(((((.......)))))..(((((.((((....-))))))))).........)))))))))(((....((......))....))). ( -31.00) >Bsubt.0 636660 119 + 4214630/2800-2920 AUGUGUCGCCACACUUCCACCUCAGACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUGC-GGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCA ..((((....)))).(((.((((((.........(((((.......)))))..(((((.((((....-))))))))).........)))))))))(((....((......))....))). ( -31.40) >Blich.0 613689 120 + 4222334/2800-2920 AUGUGUCGCCACACUUCCACCUCAGACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUAUAGGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCA ..((((....)))).(((.((((((.........(((((.......)))))..(((((.((((.....))))))))).........)))))))))(((....((......))....))). ( -31.70) >consensus ACAUGUCACCACACUUCCACCUCAAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUACCUUAC_GCAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCA .((((....))))..(((.((((((.........(((((.......)))))..(((((.((((.....))))))))).........)))))))))(((....((......))....))). (-33.35 = -30.85 + -2.50)

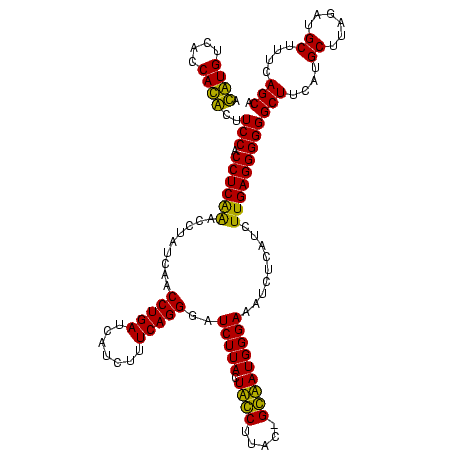
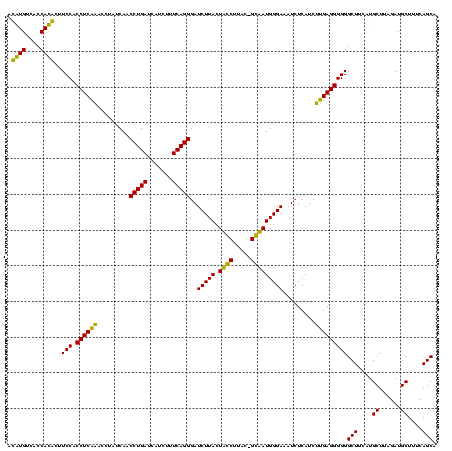
| Location | 247,889 – 248,005 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.55 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -32.88 |
| Energy contribution | -31.38 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.988457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 247889 116 + 5227293/2840-2960 AUUUCCCAUAGC-GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC---A ...((((.((((-....))))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).))).......---. ( -35.50) >Bcere.0 281279 116 + 5224283/2840-2960 AUUUCCCAUAGC-GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC---A ...((((.((((-....))))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).))).......---. ( -35.50) >Bsubt.0 636660 116 + 4214630/2840-2960 AUUUCCCAUUCC-GCAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACC---A ...((((.((((-....))))....(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))).))).......---. ( -36.60) >Blich.0 613689 120 + 4222334/2840-2960 AUUUCCCAUUCCUAUAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACCUAAA ...((((.((((.....))))....(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))).)))........... ( -33.10) >consensus AUUUCCCAUACC_GUAAGCAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCCGAGGUGGAAGCAUGGCGACACAUGGAGCUGACAAAUACUAAUAGAUCGAGGACUUAACC___A ...((((.((((.....))))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).)))........... (-32.88 = -31.38 + -1.50)

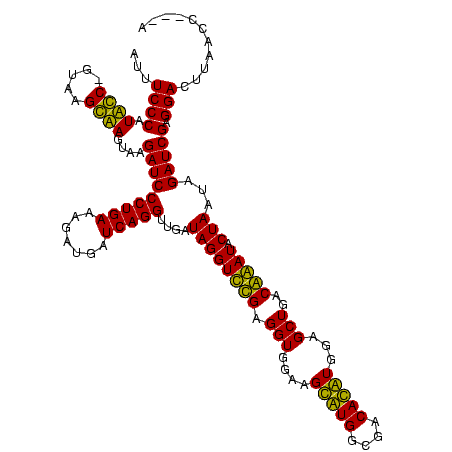
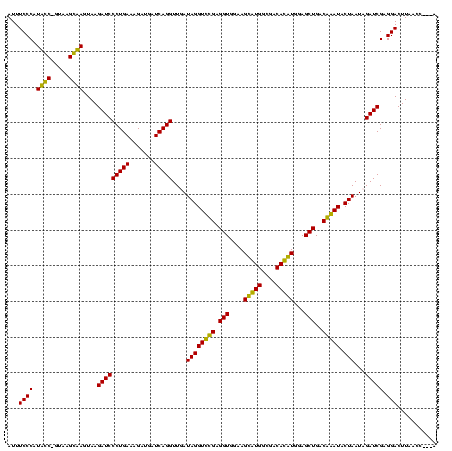
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:24:20 2006