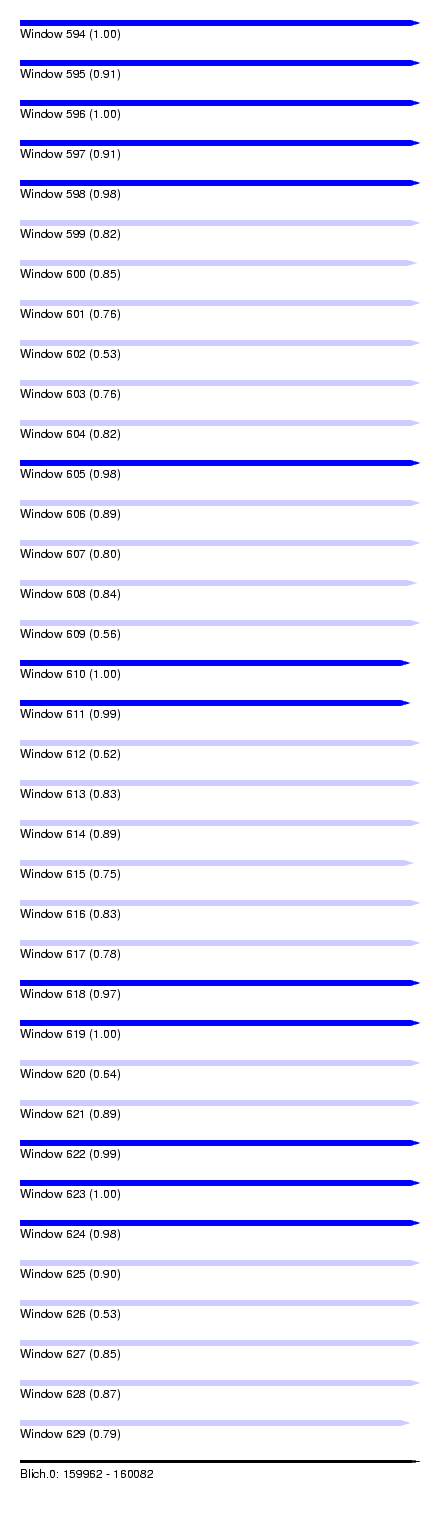
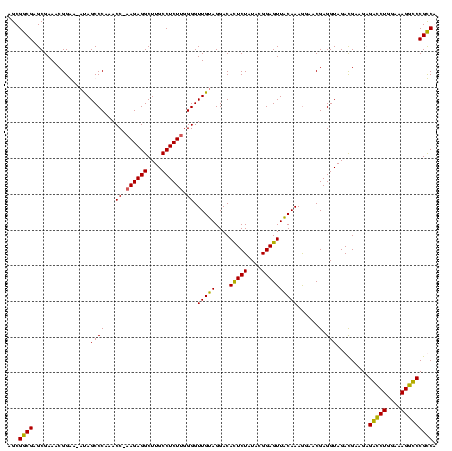
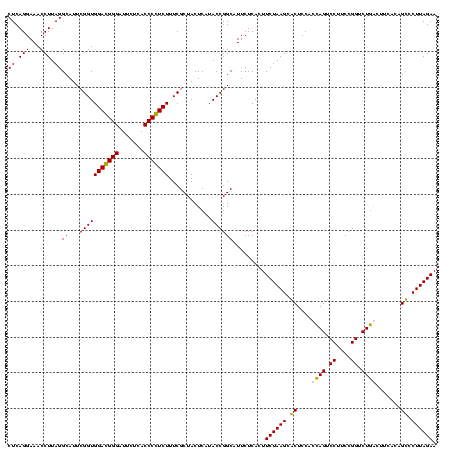
| Sequence ID | Blich.0 |
|---|---|
| Location | 159,962 – 160,082 |
| Length | 120 |
| Max. P | 0.999325 |

| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -42.25 |
| Energy contribution | -40.25 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.85 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/80-200 CGAUGAAGGACGGGACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCA .......((((((........))).....((((((((((((.......)))))...)))))))...)))(((((((....)))(((((((......)))))))............)))). ( -42.30) >Bsubt.0 162555 120 + 4214630/80-200 CGAUGAAGGACGGGACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCA .......((((((........))).....((((((((((((.......)))))...)))))))...)))(((((((....)))(((((((......)))))))............)))). ( -42.30) >Bcere.0 332443 120 + 5224283/80-200 CGAUGAAGGACGGGACUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACA .(((((.(((((((((....)........((((((((((((.......)))))...)))))))..)))))...(((....)))(((((((......))))))).))))))))........ ( -42.10) >Banth.0 291724 120 + 5227293/80-200 CGAUGAAGGACGGGACUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACA .(((((.(((((((((....)........((((((((((((.......)))))...)))))))..)))))...(((....)))(((((((......))))))).))))))))........ ( -42.10) >consensus CGAUGAAGGACGGGACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCACACGUAAUGGAAUGGUAUCCAUAUCUGAAUACA .((((..((((((........))).....((((((((((((.......)))))...)))))))...)))....(((....)))(((((((......))))))).....))))........ (-42.25 = -40.25 + -2.00)

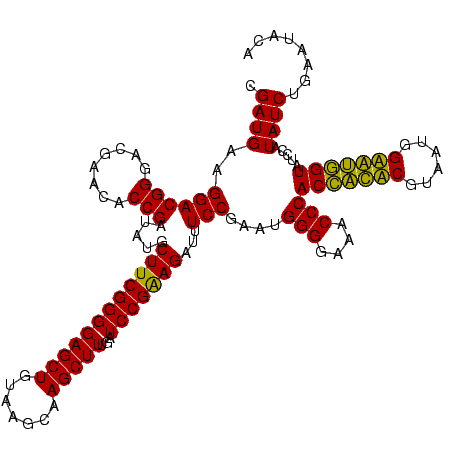
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -33.70 |
| Consensus MFE | -34.20 |
| Energy contribution | -32.95 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.60 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.909403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/80-200 UGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUUCGUCCCGUCCUUCAUCG .((((...((((((...(((((((......)))))))(((.......(((((((....)))))))...((((.......)))).)))....))))))...))))................ ( -33.70) >Bsubt.0 162555 120 + 4214630/80-200 UGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUUCGUCCCGUCCUUCAUCG .((((...((((((...(((((((......)))))))(((.......(((((((....)))))))...((((.......)))).)))....))))))...))))................ ( -33.70) >Bcere.0 332443 120 + 5224283/80-200 UGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUAGUCCCGUCCUUCAUCG ........((((((((((((((((......)))))))((((((((......))))).((((((.....((((.......))))..))))))...............))))))))).))). ( -33.70) >Banth.0 291724 120 + 5227293/80-200 UGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUAGUCCCGUCCUUCAUCG ........((((((((((((((((......)))))))((((((((......))))).((((((.....((((.......))))..))))))...............))))))))).))). ( -33.70) >consensus UGAAUUCAGAUAAGGAUACCACACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUAGUCCCGUCCUUCAUCG ........((((((((((((((((......)))))))((((((((......))))))))..((((...((((.......))))..((((......)))).....)))).)))))).))). (-34.20 = -32.95 + -1.25)

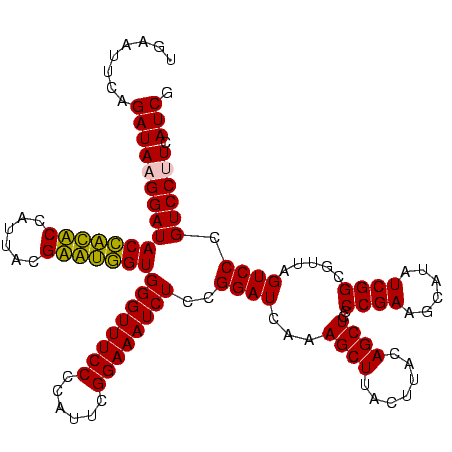

| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -35.30 |
| Consensus MFE | -35.90 |
| Energy contribution | -32.65 |
| Covariance contribution | -3.25 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.32 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996292 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/120-240 UGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUA (((..((...(((..((((...((.(((.((..(((....)))(((((((......))))))).......)).))).))...))))..)))...)).....(((....)))..))).... ( -37.20) >Bsubt.0 162555 120 + 4214630/120-240 UGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUA (((..((...(((..((((...((.(((.((..(((....)))(((((((......))))))).......)).))).))...))))..)))...)).....(((....)))..))).... ( -37.20) >Bcere.0 332443 119 + 5224283/120-240 UGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAACAUCUA (((.(((...(((((...)))))..((((....(((....)))(((((((......))))))).(((((((((.........)))))-))))...........)))))))...))).... ( -33.40) >Banth.0 291724 119 + 5227293/120-240 UGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAACAUCUA (((.(((...(((((...)))))..((((....(((....)))(((((((......))))))).(((((((((.........)))))-))))...........)))))))...))).... ( -33.40) >consensus UGUAAGCAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCACACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUA_AAGAAGACAGACCCAGGGAACUGAAACAUCUA (((..((...(((..((((......(((.((..(((....)))(((((((......))))))).......)).)))......))))..)))...)).....(((....)))..))).... (-35.90 = -32.65 + -3.25)

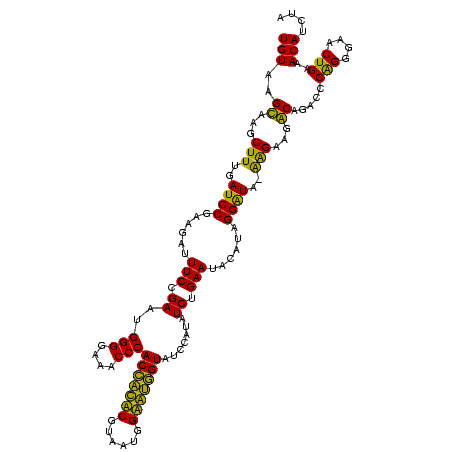
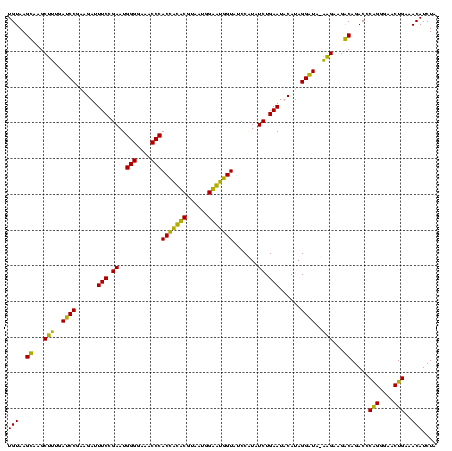
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -28.75 |
| Energy contribution | -27.25 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.911878 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/120-240 UAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACA .....((...((((....(((...(((.((.((((((...((....))))))))))((((((((......)))))))))))...)))(((((((....)))))))...)))).))..... ( -33.10) >Bsubt.0 162555 120 + 4214630/120-240 UAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACA .....((...((((....(((...(((.((.((((((...((....))))))))))((((((((......)))))))))))...)))(((((((....)))))))...)))).))..... ( -33.10) >Bcere.0 332443 119 + 5224283/120-240 UAGAUGUUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACA .....(((((((....)))((((((...((((-((..((........)).))))))((((((((......))))))))(((((((......)))))))..)))))).))))......... ( -28.70) >Banth.0 291724 119 + 5227293/120-240 UAGAUGUUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACA .....(((((((....)))((((((...((((-((..((........)).))))))((((((((......))))))))(((((((......)))))))..)))))).))))......... ( -28.70) >consensus UAGAUGUUUCAGUUCCCCGGGUCUGCCUUCCC_UACCCUAUGAAUUCAGAUAAGGAUACCACACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUACUUACA .....((((..((((.((..(((((..(((...........)))..)))))..)).((((((((......))))))))(((((((......)))))))...))))..))))......... (-28.75 = -27.25 + -1.50)

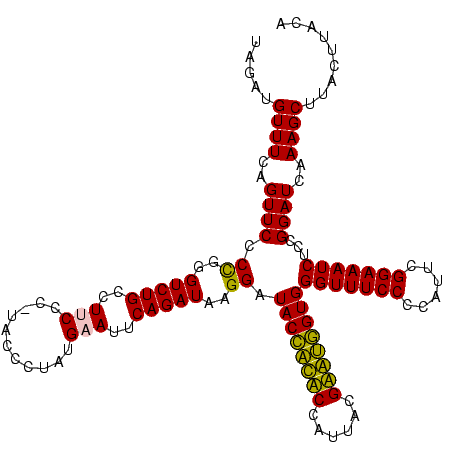
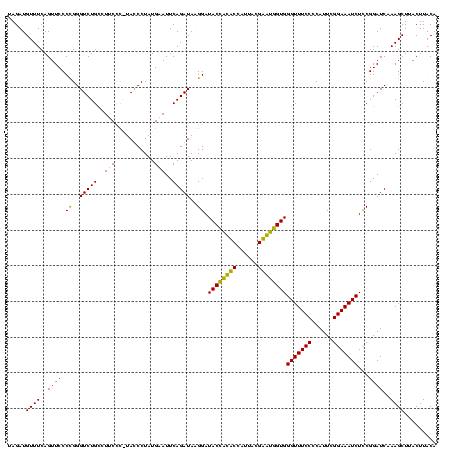
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -31.30 |
| Energy contribution | -29.30 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984127 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/160-280 CCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUCCCUGAGU .(((((((((......))))))).(((((((((.........))))))).)).))(((..(((((..(((..........))).))))).((((......((....))..)))))))... ( -37.10) >Bsubt.0 162555 120 + 4214630/160-280 CCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUCCCUGAGU .(((((((((......))))))).(((((((((.........))))))).)).))(((..(((((..(((..........))).))))).((((......((....))..)))))))... ( -37.10) >Bcere.0 332443 119 + 5224283/160-280 CCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAGAGAAAGCAAAUGCGAUUUCCUGAGU ...(((((((......))))))).(((((((((.........)))))-))))...(((..(((((..(((..........))).))))).....((((..((....))..)))))))... ( -35.00) >Banth.0 291724 119 + 5227293/160-280 CCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUA-AGGAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAGAGAAAGCAAAUGCGAUUUCCUGAGU ...(((((((......))))))).(((((((((.........)))))-))))...(((..(((((..(((..........))).))))).....((((..((....))..)))))))... ( -35.00) >consensus CCCACCACACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUA_AAGAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGCGAUUCCCUGAGU ...(((((((......))))))).(((...((((..(((...........)))..)))).(((((..(((..........))).))))).)))((.(((.((....))..))).)).... (-31.30 = -29.30 + -2.00)

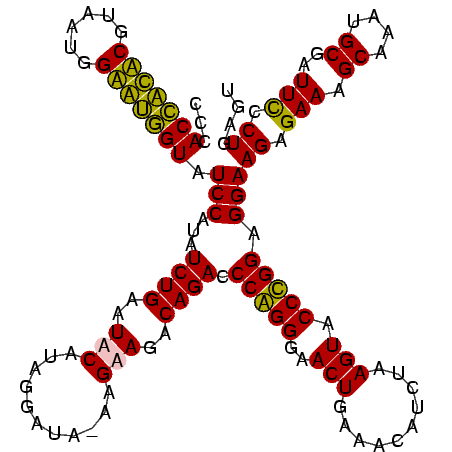
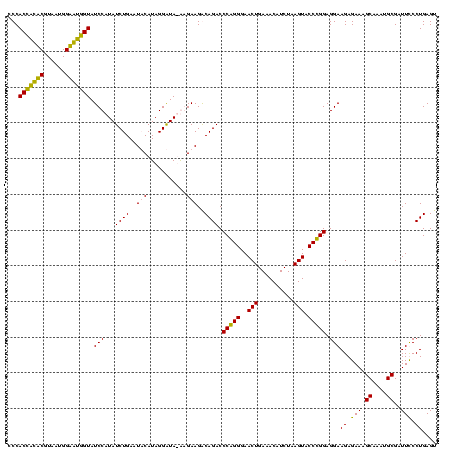
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.54 |
| Mean single sequence MFE | -31.40 |
| Consensus MFE | -31.25 |
| Energy contribution | -30.00 |
| Covariance contribution | -1.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.42 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.819141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/160-280 ACUCAGGGAAUCGCAUUUGCUUUCUCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGG ....((((((..((....)).))))))(((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..)))((((((((......)))))))).. ( -35.80) >Bsubt.0 162555 120 + 4214630/160-280 ACUCAGGGAAUCGCAUUUGCUUUCUCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGG ....((((((..((....)).))))))(((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..)))((((((((......)))))))).. ( -35.80) >Bcere.0 332443 119 + 5224283/160-280 ACUCAGGAAAUCGCAUUUGCUUUCUCUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGG ....(((((..(((....))..........((((((.(((..((....)))))..))))))...)..)))))-..(((...((((((......))))))...(((((.....)))))))) ( -27.00) >Banth.0 291724 119 + 5227293/160-280 ACUCAGGAAAUCGCAUUUGCUUUCUCUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUCCU-UACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGG ....(((((..(((....))..........((((((.(((..((....)))))..))))))...)..)))))-..(((...((((((......))))))...(((((.....)))))))) ( -27.00) >consensus ACUCAGGAAAUCGCAUUUGCUUUCUCUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCCC_UACCCUAUGAAUUCAGAUAAGGAUACCACACCAUUACGAAUGGUGGG ....((((((..((....)).))))))(((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..)))((((((((......)))))))).. (-31.25 = -30.00 + -1.25)

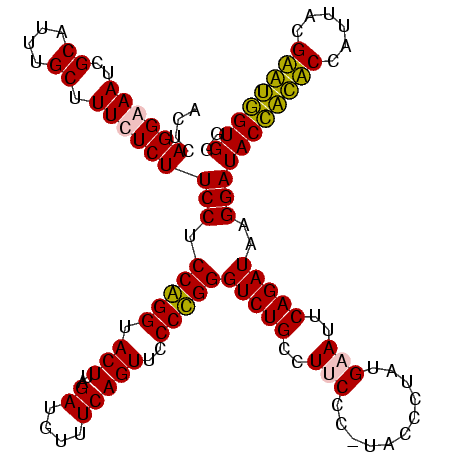
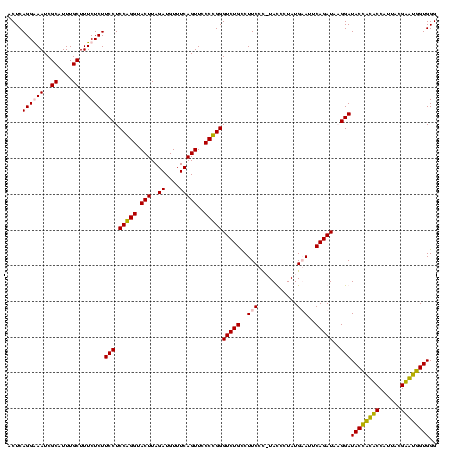
| Location | 159,962 – 160,081 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.34 |
| Mean single sequence MFE | -40.55 |
| Consensus MFE | -34.06 |
| Energy contribution | -34.38 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848573 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 119 + 4222334/280-400 AGCGGCGAGCGAAACGGGA-ACAGCCCAAACCCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUGUACGGAGUUGCAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCA ...((((.((((((((((.-....)))...(((((((((....))))))))))))........((((....))))))))......................(((((....))))))))). ( -45.90) >Bsubt.0 162555 117 + 4214630/280-400 AGCGGCGAGCGAAACGGGA-UUAGCCCAAACC-AAGAGGCUUGCCUCU-GGGGUUGUAGGACACUCUGUACGGAGUUACAAAGGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCA ...((((..((....(((.-....)))...((-.(((((....)))))-.)).(((((....(((((....)))))))))).....)).............(((((....))))))))). ( -37.70) >Bcere.0 332443 119 + 5224283/280-400 AGCGGCGAGCGAAACGGAACAUAGCCCAAACC-AAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCG ..(((((.((....((....(((((((....(-((((((....)))))))))))))).......(((((.....(((.......)))...)))))))..))(((((....)))))))))) ( -39.30) >Banth.0 291724 119 + 5227293/280-400 AGCGGCGAGCGAAACGGAACAUAGCCCAAACC-AAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCG ..(((((.((....((....(((((((....(-((((((....)))))))))))))).......(((((.....(((.......)))...)))))))..))(((((....)))))))))) ( -39.30) >consensus AGCGGCGAGCGAAACGGAA_AUAGCCCAAACC_AAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGAGACCUGGAAAGGCCCGCCA ...((((................((((...((.((((((....)))))).)).(((((....(((((....))))))))))......).))).........(((((....))))))))). (-34.06 = -34.38 + 0.31)


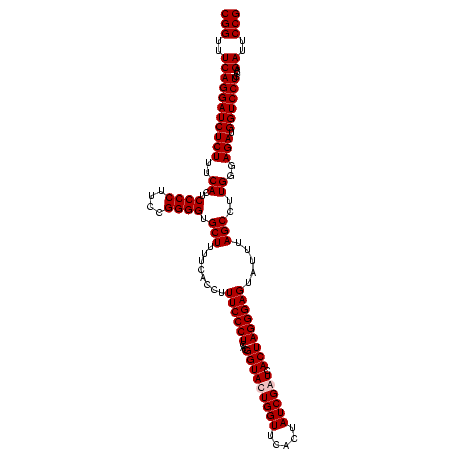
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.89 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -37.57 |
| Energy contribution | -38.07 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756135 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/480-600 CGGUUUCAGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGGUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCGGAUUCCG (((..(((((((((((..((..((((....)))).(((........((((((...(((((((((.....)))))).))))))))).....)))..))..))).))))))....))..))) ( -40.22) >Bsubt.0 162555 120 + 4214630/480-600 CGGUUUCAGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGGUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCGGAUUCCG (((..(((((((((((..((..((((....)))).(((........((((((...(((((((((.....)))))).))))))))).....)))..))..))).))))))....))..))) ( -40.22) >Bcere.0 332443 120 + 5224283/480-600 CGGUUUCAGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGAUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCAGAUUCCG (((..(((((((((((..((..((((....)))).(((........((((((...(((..(((....)))...)))...)))))).....)))..))..))).))))))....))..))) ( -35.82) >Banth.0 291724 120 + 5227293/480-600 CGGUUUCAGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGAUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCAGAUUCCG (((..(((((((((((..((..((((....)))).(((........((((((...(((..(((....)))...)))...)))))).....)))..))..))).))))))....))..))) ( -35.82) >consensus CGGUUUCAGGAUCUCUUUCACUCCCCUUCCGGGGUGCUUUUCACCUUUCCCUCACGGUACUGGUUCACUAUCGAUCACUAGGGAGUAUUUAGCCUUGGGAGAUGGUCCUCCCAGAUUCCG (((..(((((((((((..((..((((....)))).(((........((((((...(((((((((.....)))))).))))))))).....)))..))..))).))))))....))..))) (-37.57 = -38.07 + 0.50)


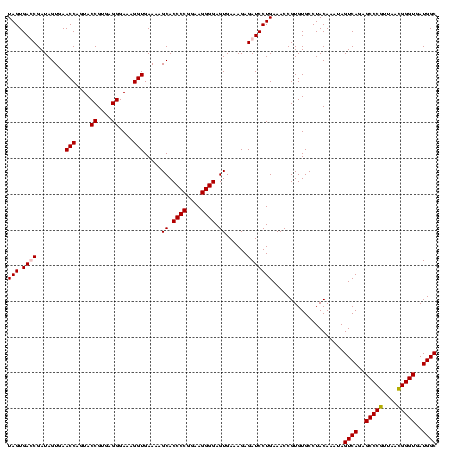
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.47 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -33.81 |
| Energy contribution | -33.62 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.90 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530913 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/520-640 UAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGC (((..((((.(((.(((((....((....))....))).....((.((((....)))).))........)))))....)).))..)))......((((..(((((....)))))..)))) ( -37.10) >Bsubt.0 162555 120 + 4214630/520-640 UAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGC (((..((((.(((.(((((....((....))....))).....((.((((....)))).))........)))))....)).))..)))......((((..(((((....)))))..)))) ( -37.10) >Bcere.0 332443 120 + 5224283/520-640 UAGUGAUCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGC (((.((((........(((....((....))....))).....((.((((....)))).))......)))))))....................((((..(((((....)))))..)))) ( -39.50) >Banth.0 291724 120 + 5227293/520-640 UAGUGAUCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAAUGGGUGAUGGC (((.((((........(((....((....))....))).....((.((((....)))).))......)))))))....................((((..(((((....)))))..)))) ( -36.90) >consensus UAGUGACCGAUAGUGAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGC ....................((((((...((((..........((.((((....)))).))........)))).....)).)))).........((((..(((((....)))))..)))) (-33.81 = -33.62 + -0.19)

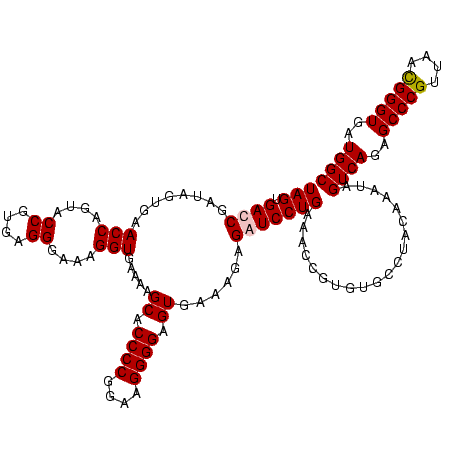
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.61 |
| Mean single sequence MFE | -42.75 |
| Consensus MFE | -41.61 |
| Energy contribution | -41.17 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.761663 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/560-680 AAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUG ...((.((((....)))).))....(.((((.(((..(((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....)))))....))).)))).)... ( -42.40) >Bsubt.0 162555 120 + 4214630/560-680 AAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCUCGUG ...((.((((....)))).))....((((((.(((..(((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....)))))....))).))))))... ( -47.20) >Bcere.0 332443 120 + 5224283/560-680 AAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUG ...((.((((....)))).))....(.((((.(((..(((.((..(((((((..((((..(((((....)))))..))))......))))))).....)))))....))).)))).)... ( -42.00) >Banth.0 291724 120 + 5227293/560-680 AAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUG ...((.((((....)))).))....(.((((.(((..(((.((..(((((((..((((..(((((....)))))..))))......))))))).....)))))....))).)))).)... ( -39.40) >consensus AAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUG ...((.((((....)))).))....(.((((.(((..(((.((..(((((((..((((..(((((....)))))..))))......))))))).....)))))....))).)))).)... (-41.61 = -41.17 + -0.44)


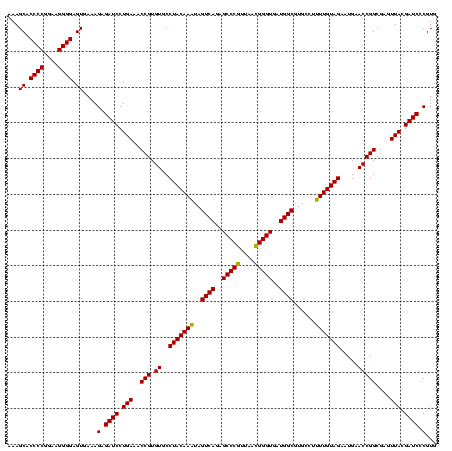
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -40.17 |
| Energy contribution | -38.55 |
| Covariance contribution | -1.62 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/600-720 UGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGUUUGAAAAGACGGAGCCGCAGCGAAAGCGA ..((((((((((((((((..(((((....)))))..)))).)))...))))))..((...(((.((.......)))))..)).((((..((((....))))..)))))))((....)).. ( -42.50) >Bsubt.0 162555 119 + 4214630/600-720 UGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCUCGUGCAAGGUUAAGCA-GAAGAUGCGGAGCCGCAGCGAAAGCGA (.((((((((((((((((..(((((....)))))..)))).)))...))))))....((((.(((((.......)))))....))))..)))-.)...((((....))))((....)).. ( -43.90) >Bcere.0 332443 119 + 5224283/600-720 UGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGA ((((.((.(.....((((..(((((....)))))..))))..(((((((.(((....((((..((...((((....)))))).))))...))-))))))))).)).))))((....)).. ( -43.40) >Banth.0 291724 119 + 5227293/600-720 UGUGCCUACAAAUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGA ((((.((.(.....((((..(((((....)))))..))))..(((((((.(((....((((..((...((((....)))))).))))...))-))))))))).)).))))((....)).. ( -40.80) >consensus UGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGCU_GAAGAGGCGGAGCCGCAGCGAAAGCGA ..((((((((((..((((..(((((....)))))..))))......)))))))..((...(((.((.......)))))..)).((((..(((......)))..)))))))((....)).. (-40.17 = -38.55 + -1.62)

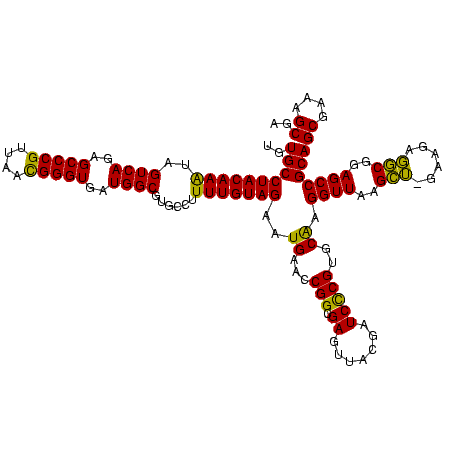
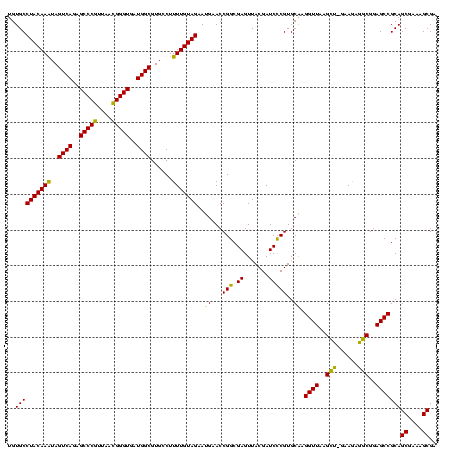
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -49.12 |
| Consensus MFE | -49.36 |
| Energy contribution | -48.68 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.63 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/760-880 ACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACC ....(((..(((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))))(((.....)))......))) ( -50.10) >Bsubt.0 162555 120 + 4214630/760-880 ACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAGCC ....(((..(((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))))(((.....)))......))) ( -50.40) >Bcere.0 332443 120 + 5224283/760-880 ACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACC ....(((...((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))).((((...))))......))) ( -48.00) >Banth.0 291724 120 + 5227293/760-880 ACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACC ....(((...((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))).((((...))))......))) ( -48.00) >consensus ACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUGCCAAUCGAACC ....(((...((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))).(((.....)))......))) (-49.36 = -48.68 + -0.69)

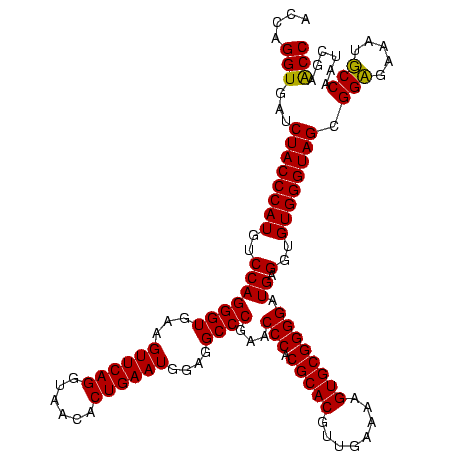
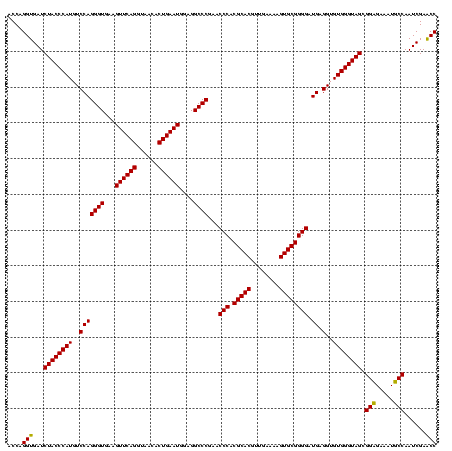
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.92 |
| Mean single sequence MFE | -38.02 |
| Consensus MFE | -36.60 |
| Energy contribution | -36.85 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/760-880 GGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGU (((..((........))....(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...))).... ( -36.80) >Bsubt.0 162555 120 + 4214630/760-880 GGCUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGU .(((.....))).........(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).))))))).......... ( -36.50) >Bcere.0 332443 120 + 5224283/760-880 GGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGU (((..(((.(((.....))).(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).))))))))))))).... ( -39.40) >Banth.0 291724 120 + 5227293/760-880 GGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGU (((..(((.(((.....))).(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).))))))))))))).... ( -39.40) >consensus GGUUCGAUUGGAAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGU (((..(((.((.......)).(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).))))))))))))).... (-36.60 = -36.85 + 0.25)

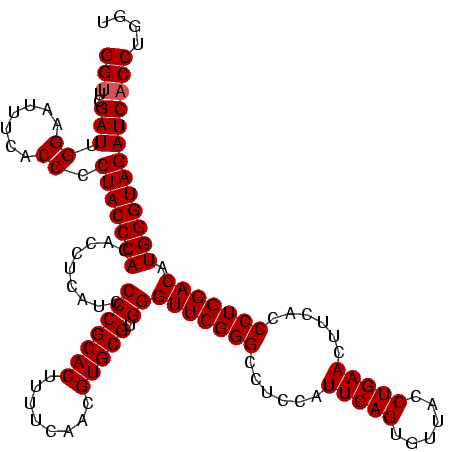
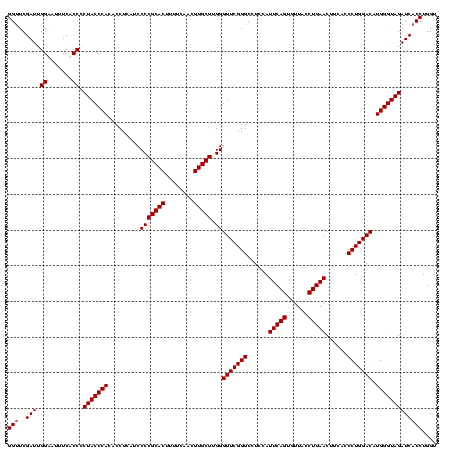
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.36 |
| Mean single sequence MFE | -35.01 |
| Consensus MFE | -33.26 |
| Energy contribution | -32.83 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.800027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/800-920 CUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUG .((((......))))....((((.(((((...........))))).............(((..((((((...........((((........))))))))))..))).))))........ ( -32.50) >Bsubt.0 162555 120 + 4214630/800-920 CUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGCUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUG .((((......))))....(((((..........)))))((((((((.(((..............)))........((((((((........))))).))).)))))))).......... ( -34.34) >Bcere.0 332443 120 + 5224283/800-920 CUAGCCCUAAAGCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUG .((((......))))....(((((((........)))))((((((((..(((.....)))................((((((((........))))).))).))))))))))........ ( -36.60) >Banth.0 291724 120 + 5227293/800-920 CUAGCCCUAAAGCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUG .((((......))))....(((((((........)))))((((((((..(((.....)))................((((((((........))))).))).))))))))))........ ( -36.60) >consensus CUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUG .((((......))))....(((((..........)))))((((((((..((.......))................((((((((........))))).))).)))))))).......... (-33.26 = -32.83 + -0.44)

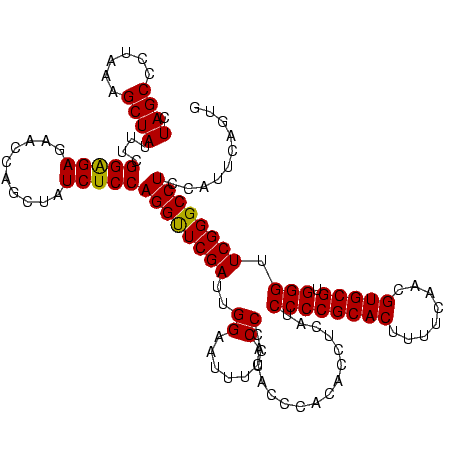
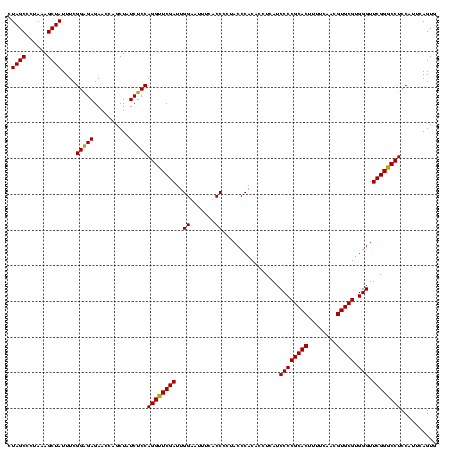
| Location | 159,962 – 160,081 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -24.80 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.18 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.839586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 119 + 4222334/840-960 UCCAAUCAGUGCUCUACCUCCAAGACUCUUAC-CUUGAGGCUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCC .((((((....((((.((((.(((........-))))))).((((......))))....)))).(((((...........)))))))))))............................. ( -25.60) >Bsubt.0 162555 119 + 4214630/840-960 UCCAAUCAGUGCUCUACCUCCAAGACUCUUAC-CUUGAGGCUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGCUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCC .((((((.(.((.((.((((.(((........-))))))).((((......))))....(((((..........))))))))).)))))))............................. ( -25.00) >Bcere.0 332443 120 + 5224283/840-960 UCCAAUCAGUGCUCUACCUCCAAGACUCUUACACUUAAGGCUAGCCCUAAAGCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCC (((((((..........((((.....(((((....))))).((((......))))....)))).(((((...........))))))))))))............................ ( -24.30) >Banth.0 291724 120 + 5227293/840-960 UCCAAUCAGUGCUCUACCUCCAAGACUCUUACACUUAAGGCUAGCCCUAAAGCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCC (((((((..........((((.....(((((....))))).((((......))))....)))).(((((...........))))))))))))............................ ( -24.30) >consensus UCCAAUCAGUGCUCUACCUCCAAGACUCUUAC_CUUAAGGCUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCACCCCUACCCACACCUCAUCCCC .((((((..........((((.....(((((....))))).((((......))))....)))).(((((...........)))))))))))............................. (-22.42 = -22.18 + -0.25)

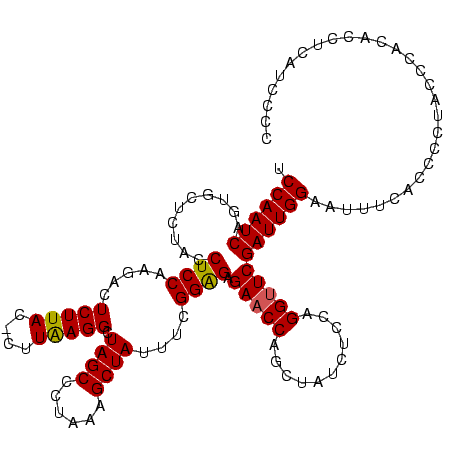
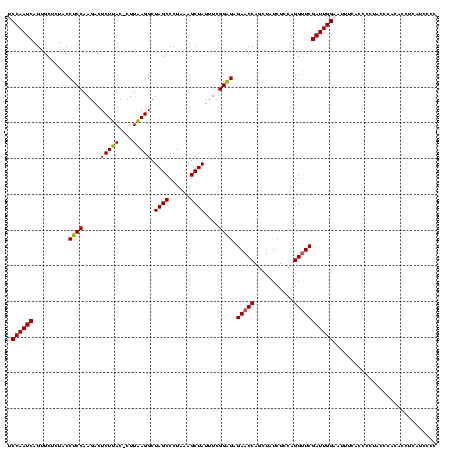
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -36.05 |
| Consensus MFE | -35.90 |
| Energy contribution | -35.40 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.564068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/1120-1240 GAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAUCACCGAA ((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..)))).((.(((.........))).))................ ( -36.40) >Bsubt.0 162555 120 + 4214630/1120-1240 GAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAUCACCGAA ((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..)))).((.(((.........))).))................ ( -36.40) >Bcere.0 332443 120 + 5224283/1120-1240 GAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAUACACCACCGAA ((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..)))).((.(((.........))).))................ ( -35.70) >Banth.0 291724 120 + 5227293/1120-1240 GAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAUACACCACCGAA ((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..)))).((.(((.........))).))................ ( -35.70) >consensus GAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACACACCACCGAA ((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..)))).((.(((.........))).))................ (-35.90 = -35.40 + -0.50)

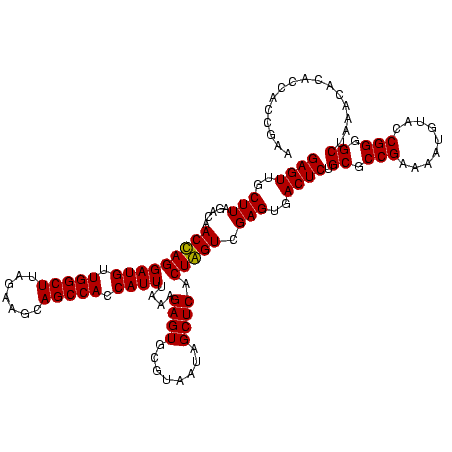
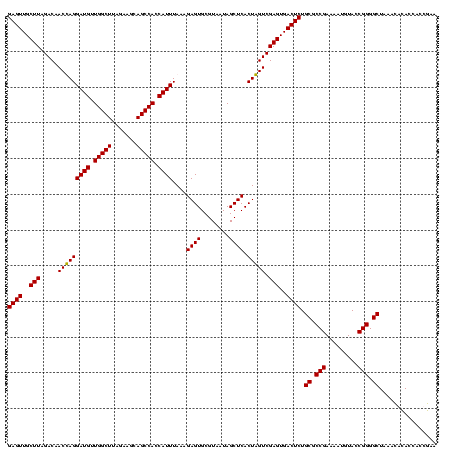
| Location | 159,962 – 160,079 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.82 |
| Mean single sequence MFE | -44.42 |
| Consensus MFE | -44.92 |
| Energy contribution | -43.05 |
| Covariance contribution | -1.87 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.51 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998128 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 117 + 4222334/1240-1360 GCUGCGGACUGUUCU---UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGG (((((..(((((((.---...)))))))((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))).......)))))........ ( -48.50) >Bsubt.0 162555 116 + 4214630/1240-1360 GCUGCGGACUGUUCU---U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGG (((((..(((((((.---.-.)))))))((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))).......)))))........ ( -47.60) >Bcere.0 332443 120 + 5224283/1240-1360 GCUGCGGAUUGAUACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACG (((((..(((((((((...)))))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))).......)))))........ ( -40.80) >Banth.0 291724 120 + 5227293/1240-1360 GCUGCGGAUUGAUACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACG (((((..(((((((((...)))))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))).......)))))........ ( -40.80) >consensus GCUGCGGACUGAUAC___UGCGAACAGUGGUAGGAGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACG (((((..((((((((.....))))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))).......)))))........ (-44.92 = -43.05 + -1.87)

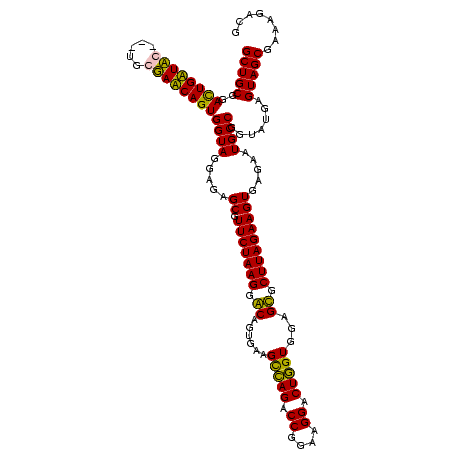
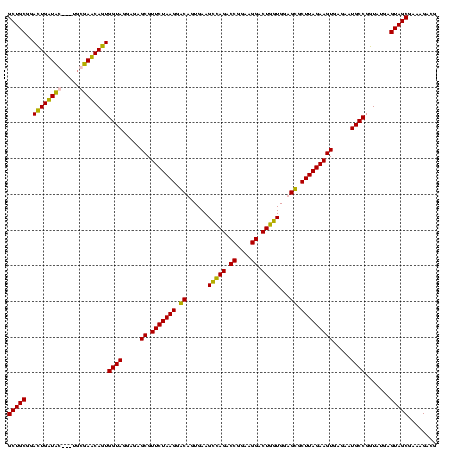
| Location | 159,962 – 160,079 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.82 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -22.83 |
| Energy contribution | -22.46 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 117 + 4222334/1240-1360 CCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCGCA---AGAACAGUCCGCAGC ........((..........((.........(((((((.(((...((((.((....)).))))......))).)))))))..........))(((((((...---.)))))))..))... ( -31.41) >Bsubt.0 162555 116 + 4214630/1240-1360 CCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCG-A---AGAACAGUCCGCAGC ........((..........((.........(((((((.(((...((((.((....)).))))......))).)))))))..........))(((((((.-.---.)))))))..))... ( -26.71) >Bcere.0 332443 120 + 5224283/1240-1360 CGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGGUAUCAAUCCGCAGC ........((..........((.........(((((((.((.....(((.((....)).)))........)).)))))))..........))..(((((((...)))))))....))... ( -21.53) >Banth.0 291724 120 + 5227293/1240-1360 CGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGGUAUCAAUCCGCAGC ........((..........((.........(((((((.((.....(((.((....)).)))........)).)))))))..........))..(((((((...)))))))....))... ( -21.53) >consensus CCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACAGCCCUUAGAACGCUCCCCUACCACUGAUACCA___AGAACAAUCCGCAGC ........((..........((.........(((((((.((....((((.((....)).)))).......)).)))))))..........))((((((((.....))))))))..))... (-22.83 = -22.46 + -0.37)

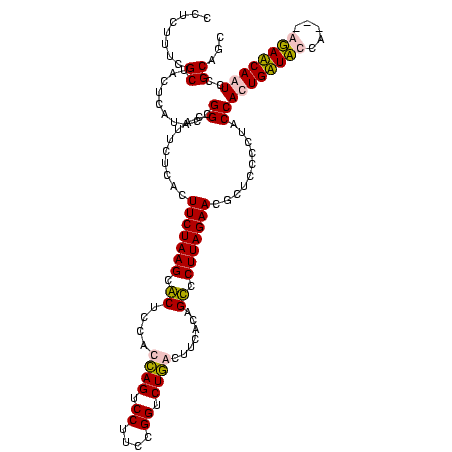
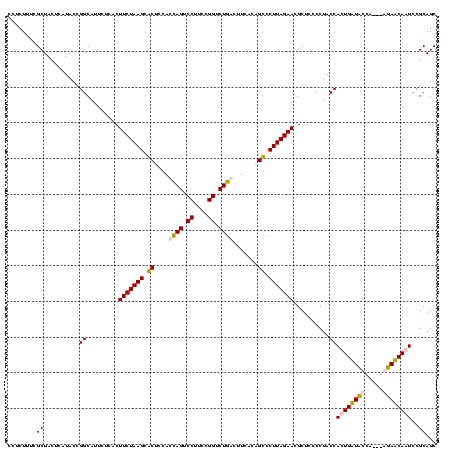
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -39.10 |
| Consensus MFE | -39.83 |
| Energy contribution | -38.20 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.40 |
| Structure conservation index | 1.02 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.615799 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/1280-1400 UUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGGGGUGAGAAUCCCCUCCACCGAAUGCCUAAGGUUUCCUGAG (((((((.(((.(...(((((.((....)).)))))).))).)))))))....(((.((((((((......((...((((((.......))))))...)).)))))...))))))..... ( -43.70) >Bsubt.0 162555 120 + 4214630/1280-1400 UUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGGGGUGAGAAUCCCCUCCACCGAAUGCCUAAGGUUUCCUGAG (((((((.(((.(...(((((.((....)).)))))).))).)))))))....(((.((((((((......((...((((((.......))))))...)).)))))...))))))..... ( -44.30) >Bcere.0 332443 120 + 5224283/1280-1400 UUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAAUGCCUAAGGUUUCCUGAG (((((((.((......(((((.((....)).)))))...)).)))))))....(((.((((((((......((...((((((.......))))))...)).)))))...))))))..... ( -34.20) >Banth.0 291724 120 + 5227293/1280-1400 UUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAAUGCCUAAGGUUUCCUGAG (((((((.((......(((((.((....)).)))))...)).)))))))....(((.((((((((......((...((((((.......))))))...)).)))))...))))))..... ( -34.20) >consensus UUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCCUCCACCGAAUGCCUAAGGUUUCCUGAG (((((((.((......(((((.((....)).)))))...)).)))))))....(((.((((((((......((...((((((.......))))))...)).)))))...))))))..... (-39.83 = -38.20 + -1.63)

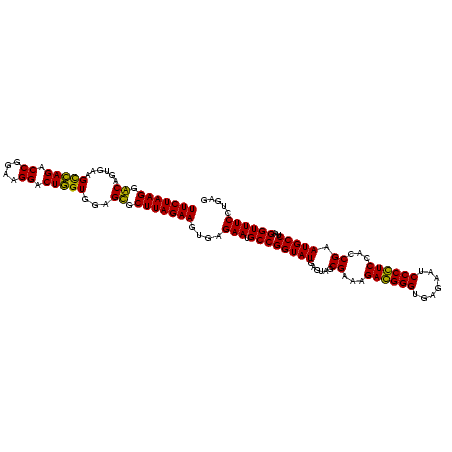
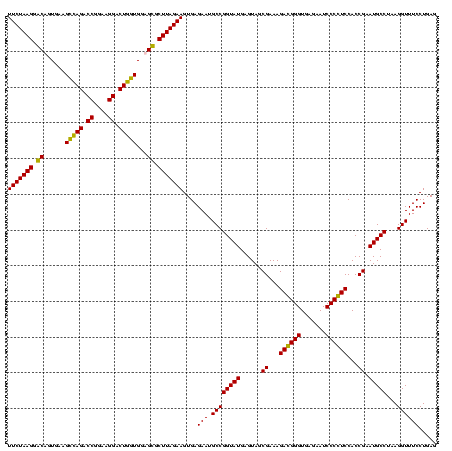
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.83 |
| Mean single sequence MFE | -31.26 |
| Consensus MFE | -29.88 |
| Energy contribution | -29.00 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/1280-1400 CUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAA ((.(((...))).))((...(((((((((((.......)))))))............)))))).........((((((.(((...((((.((....)).))))......))).)))))). ( -33.20) >Bsubt.0 162555 120 + 4214630/1280-1400 CUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAA ((.(((...))).))((...(((((((((((.......)))))))............)))))).........((((((.(((...((((.((....)).))))......))).)))))). ( -33.80) >Bcere.0 332443 120 + 5224283/1280-1400 CUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAA ....(....)((.(((....(((((((((((.......)))))).........(((.(((((.............(((.(((.....))).)))))))).)))...))))).))).)).. ( -29.01) >Banth.0 291724 120 + 5227293/1280-1400 CUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAA ....(....)((.(((....(((((((((((.......)))))).........(((.(((((.............(((.(((.....))).)))))))).)))...))))).))).)).. ( -29.01) >consensus CUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACAGCCCUUAGAA ((.(((...))).))((...(((((((((((.......)))))))............)))))).........((((((.((....((((.((....)).)))).......)).)))))). (-29.88 = -29.00 + -0.88)

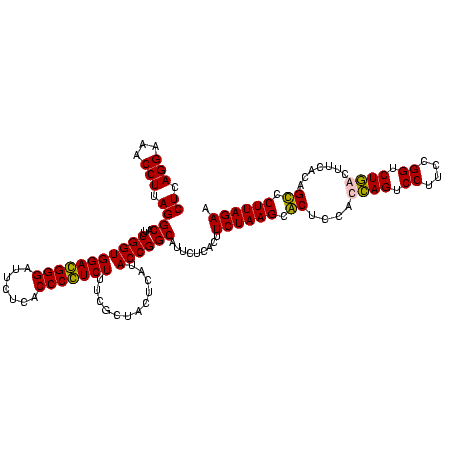
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.56 |
| Mean single sequence MFE | -35.48 |
| Consensus MFE | -32.31 |
| Energy contribution | -30.81 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.96 |
| SVM RNA-class probability | 0.890671 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/1440-1560 CUUGGACGCGGAUAUCCAAUACCGCGCUUACCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAAUGGUGAGGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUUUCGGCC ...((((((((..........))))((...............)))))).......(((.(((.(((((((.(((((.(((((........)))))...))))).))).))))))).))). ( -39.76) >Bsubt.0 162555 120 + 4214630/1440-1560 CUUGGACGCGGAUAUCCAAUACCGCGCUUACCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAAUGGUGAGGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUUUCGGCC ...((((((((..........))))((...............)))))).......(((.(((.(((((((.(((((.(((((........)))))...))))).))).))))))).))). ( -39.76) >Bcere.0 332443 120 + 5224283/1440-1560 CUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUGUCGGCC ..(((((((((..........))))(((((((.((((.....(((.........)))......)))))))))))...(((((........))))).)))))..(((.((....)).))). ( -31.20) >Banth.0 291724 120 + 5227293/1440-1560 CUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUGUCGGCC ..(((((((((..........))))(((((((.((((.....(((.........)))......)))))))))))...(((((........))))).)))))..(((.((....)).))). ( -31.20) >consensus CUUGGACGCGCACAACCAAUACCACGCUUACCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCCUACGCCUGUCGGCC ...((((((((..........))))((...............))))))((.(((((......))))).)).(((((.(((((........)))))...)))))......(((....))). (-32.31 = -30.81 + -1.50)

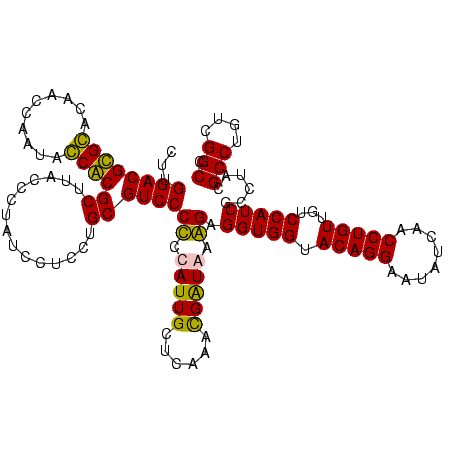
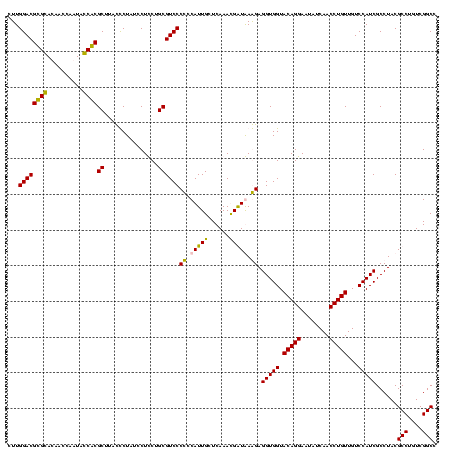
| Location | 159,962 – 160,080 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.52 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -33.50 |
| Energy contribution | -30.75 |
| Covariance contribution | -2.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753043 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 118 + 4222334/1520-1640 GAGGAUAGGGUAAGCGCGGUAUUGGAUAUCCGCGUCCAAGCAGUUAGGCUGGGAAAUAGGCAAAUCCGUUUCCCGU-AAGGCUGAGCUGUGAUGGCGA-GCGAAAUAUAGUAGCGAAGUU ..............(((.(((((((....))((..(((.((((((.(((((((((((.((.....)))))))))..-..)))).))))))..)))...-))..)))))....)))..... ( -34.10) >Bsubt.0 162555 118 + 4214630/1520-1640 GAGGAUAGGGUAAGCGCGGUAUUGGAUAUCCGCGUCCAAGCAGUUAGGCUGGGAAAUAGGCAAAUCCGUUUCCCAU-AAGGCUGAGCUGUGAUGGCGA-GCGAAAUAUAGUAGCGAAGUU ..............(((.(((((((....))((..(((.((((((.(((((((((((.((.....)))))))))..-..)))).))))))..)))...-))..)))))....)))..... ( -34.10) >Bcere.0 332443 119 + 5224283/1520-1640 AAGGAUAGAAGAAGCGUGCGAUUGGUUGUGCACGUCCAAGCAGUUAGGCUGAUAAGUAGGCAAAUCCGCUUAUCGUGAAGGCUGAGCUGUGAUGGGGAAGCUCCUUAUGG-AGCGAAGUC .............(((((((........)))))))(((.((((((.(((((((((((.((.....))))))))).....)))).))))))..)))....(((((....))-)))...... ( -39.70) >Banth.0 291724 119 + 5227293/1520-1640 AAGGAUAGAAGAAGCGUGCGAUUGGUUGUGCACGUCCAAGCAGUUAGGCUGAUAAGUAGGCAAAUCCGCUUAUCGUGAAGGCUGAGCUGUGAUGGGGAAGCUCCUUAUGG-AGCGAAGUC .............(((((((........)))))))(((.((((((.(((((((((((.((.....))))))))).....)))).))))))..)))....(((((....))-)))...... ( -39.70) >consensus AAGGAUAGAAGAAGCGCGCGAUUGGAUAUCCACGUCCAAGCAGUUAGGCUGAGAAAUAGGCAAAUCCGCUUACCGU_AAGGCUGAGCUGUGAUGGCGA_GCGAAAUAUAG_AGCGAAGUC .............((((((.((.....))))))))(((.((((((.(((((((((((.((.....))))))))).....)))).))))))..)))....((...........))...... (-33.50 = -30.75 + -2.75)

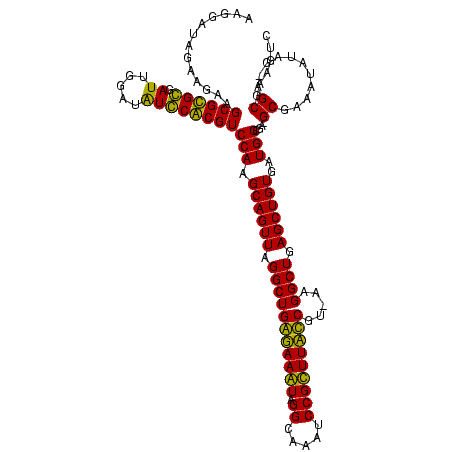
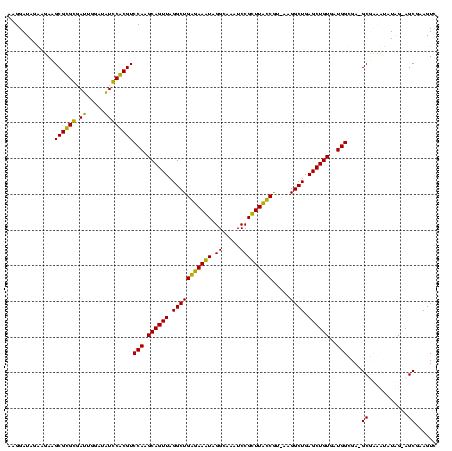
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -40.85 |
| Consensus MFE | -33.97 |
| Energy contribution | -35.60 |
| Covariance contribution | 1.63 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.829766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/1760-1880 CGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCG ((((......(((((....(....).....)))))(((((.((.((((....)))))).)))))(((((.....(((((.((....))((((.....))))...))))))))))..)))) ( -45.80) >Bsubt.0 162555 120 + 4214630/1760-1880 CGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCG ((((......(((((....(....).....)))))(((((.((.((((....)))))).)))))(((((.....(((((.((....))((((.....))))...))))))))))..)))) ( -45.80) >Bcere.0 332443 109 + 5224283/1760-1880 CGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUUUCUUAACG-----------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCG ((((......(((((....(....).....)))))(((((((....)-----------))))))((((((((((((((....))..)))))))((.........))...)))))..)))) ( -35.90) >Banth.0 291724 109 + 5227293/1760-1880 CGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUUUCUUAACG-----------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCG ((((......(((((....(....).....)))))(((((((....)-----------))))))((((((((((((((....))..)))))))((.........))...)))))..)))) ( -35.90) >consensus CGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUCUUAACG___________GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCG ((((......(((((.....(((....))))))))((((((...((((....))))..))))))(((((.((....((....))..((((...........)))).)).)))))..)))) (-33.97 = -35.60 + 1.63)

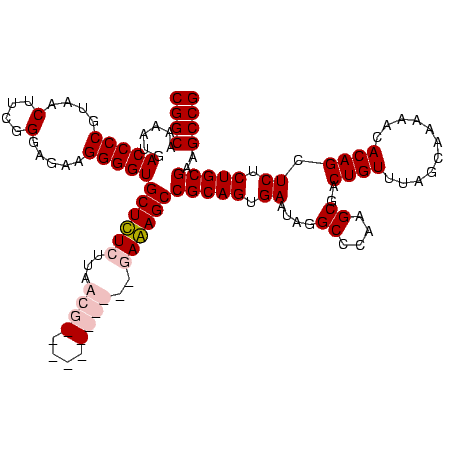
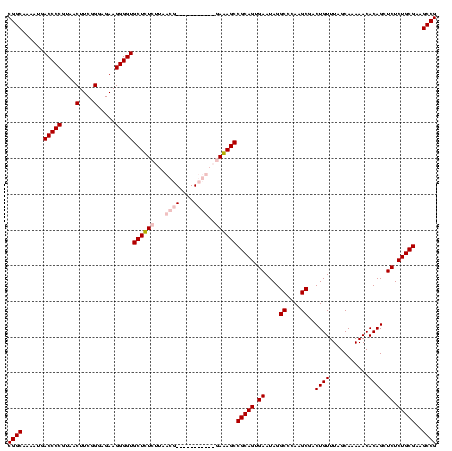
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -40.60 |
| Consensus MFE | -37.05 |
| Energy contribution | -39.05 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.780046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/1840-1960 AGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUU (((((..(((((.....)))))..(((((((((...(((....)))...))..)))))))..)))))((((..((((...(.((((((.......)))))).).))))..)).))..... ( -41.80) >Bsubt.0 162555 120 + 4214630/1840-1960 AGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUU (((((..(((((.....)))))..(((((((((...(((....)))...))..)))))))..)))))((((..((((...(.((((((.......)))))).).))))..)).))..... ( -41.80) >Bcere.0 332443 120 + 5224283/1840-1960 AAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACU ....((((.((((((.....(((..((((.(((...(((....)))...))).)))).)))..(((.....))))))))).)))).((....((((((.((....))..))))))...)) ( -39.40) >Banth.0 291724 120 + 5227293/1840-1960 AAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACU ....((((.((((((.....(((..((((.(((...(((....)))...))).)))).)))..(((.....))))))))).)))).((....((((((.((....))..))))))...)) ( -39.40) >consensus AAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAAGAGCGCUUAGCGUAAGCGAAGCUCCGAACU ....((((.((((((.........(((((((((...(((....)))...))..)))))))...(((.....))))))))).)))).......((((((.((....))..))))))..... (-37.05 = -39.05 + 2.00)

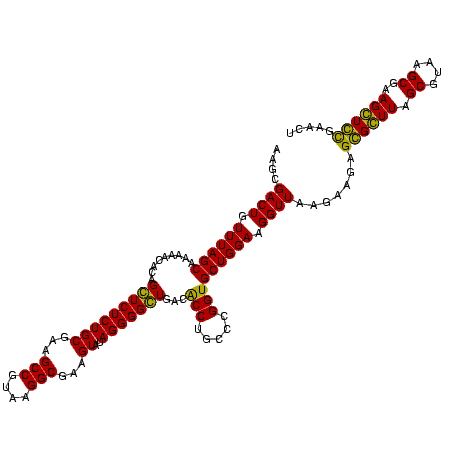
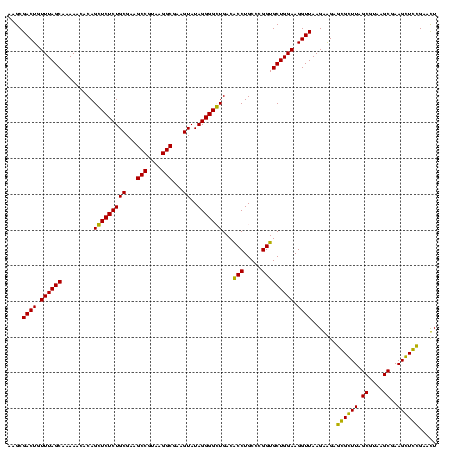
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -45.00 |
| Consensus MFE | -40.80 |
| Energy contribution | -42.05 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.91 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/1880-2000 UAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUA ..(((((.(((......)))..)))))(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))... ( -48.00) >Bsubt.0 162555 120 + 4214630/1880-2000 UAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUA ..(((((.(((......)))..)))))(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))... ( -48.00) >Bcere.0 332443 120 + 5224283/1880-2000 UAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUA ..............((((....).)))(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))... ( -42.00) >Banth.0 291724 120 + 5227293/1880-2000 UAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUA ..............((((....).)))(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))... ( -42.00) >consensus UAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAAGAGCGCUUAGCGUAAGCGAAGCUCCGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUA ..(((((.(((......)))..)))))(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))... (-40.80 = -42.05 + 1.25)

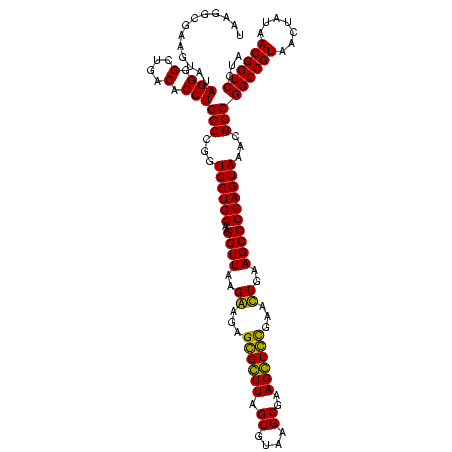
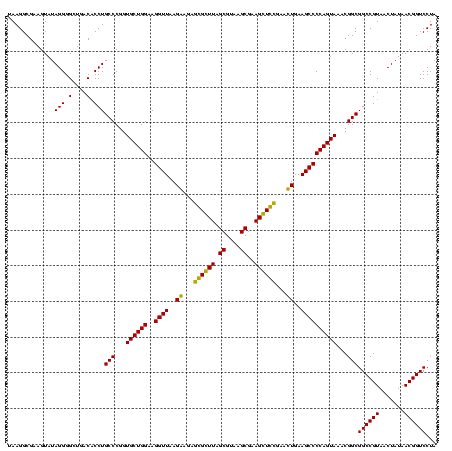
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -49.35 |
| Consensus MFE | -49.45 |
| Energy contribution | -47.95 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.37 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.51 |
| SVM RNA-class probability | 0.999325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2160-2280 AUAUUGAAUGUUGGUACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGC .....(((.((..((((((((.(..............(((((((....((((((((....)))))))).))))).))(((((....)))))).))))))))..)).)))........... ( -46.50) >Bsubt.0 162555 120 + 4214630/2160-2280 AUAUUGAAUGUUGGUACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGC .....(((.((..((((((((.(..............(((((((....((((((((....)))))))).))))).))(((((....)))))).))))))))..)).)))........... ( -46.50) >Bcere.0 332443 120 + 5224283/2160-2280 AUAUUGAAUUUUGGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGG .....((((((..(((((((.....((((.....(((..(((((....((((((((....)))))))).))))).)))((((....)))))))))))))))..))))))...((....)) ( -52.20) >Banth.0 291724 120 + 5227293/2160-2280 AUAUUGAAUUUUGGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGG .....((((((..(((((((.....((((.....(((..(((((....((((((((....)))))))).))))).)))((((....)))))))))))))))..))))))...((....)) ( -52.20) >consensus AUAUUGAAUGUUGGUACAGCUUGUACAGGAUAGGCAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCACCGGUGGGAUACCACCCUGACUGUAUUGAAAUUCUAACCCACGGC .....((((((..(((((((.....(((........((.(((((....((((((((....)))))))).))))).))(((((....)))))))))))))))..))))))........... (-49.45 = -47.95 + -1.50)

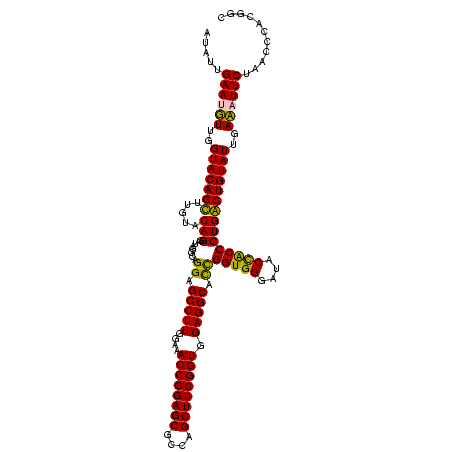

| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -29.64 |
| Energy contribution | -28.14 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.638739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2160-2280 GCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCUGUACAAGCUGUACCAACAUUCAAUAU (((((..(((((.((((......((((..((..(((((.....)))))..))...((((((((....))))))))...))))...))))...))).))..)))))............... ( -35.50) >Bsubt.0 162555 120 + 4214630/2160-2280 GCGGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCUGUACAAGCUGUACCAACAUUCAAUAU (((((..(((((.((((......((((..((..(((((.....)))))..))...((((((((....))))))))...))))...))))...))).))..)))))............... ( -34.80) >Bcere.0 332443 120 + 5224283/2160-2280 CCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGUACCAAAAUUCAAUAU ...........((((((...(((((((((((((((.....))).))((((..((((.(((....))).)))).....))))..........)))).....))))))...))))))..... ( -31.60) >Banth.0 291724 120 + 5227293/2160-2280 CCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGUACCAAAAUUCAAUAU ...........((((((...(((((((((((((((.....))).))((((..((((.(((....))).)))).....))))..........)))).....))))))...))))))..... ( -31.60) >consensus CCCGCAGGUUAGAAGGUCAAUACAGCCAGGGCAGUAUCCCACCAACGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCACCUAUCCUGUACAAACUGUACCAAAAUUCAAUAU ...........((((((...((((((((((((((............((((..((((.(((....))).)))).....))))..))))....)))).....))))))...))))))..... (-29.64 = -28.14 + -1.50)

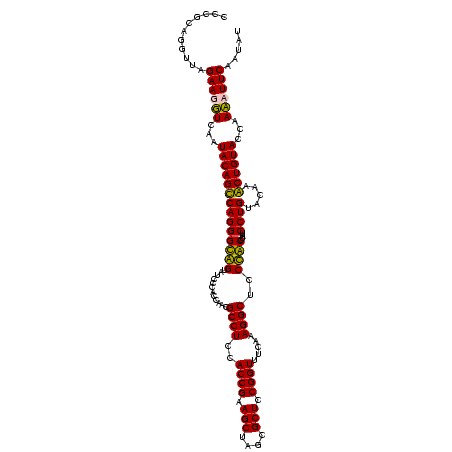
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.31 |
| Mean single sequence MFE | -48.55 |
| Consensus MFE | -47.80 |
| Energy contribution | -45.42 |
| Covariance contribution | -2.37 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.98 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888626 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2200-2320 CCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGA ((((....((((((((....)))))))).))))....(((((....)))))(..(((((.((((((((((...((.((((((.....))))))))))).)).)))))....)))))..). ( -50.30) >Bsubt.0 162555 120 + 4214630/2200-2320 CCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUUUCAGGUGGGCAGUUUGA ........((((((((....))))))))(((((((.(((((((......)))..))))...)).)))))...((((((((((.....))))..(....).......))))))........ ( -48.70) >Bcere.0 332443 120 + 5224283/2200-2320 CCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGA ((((....((((((((....)))))))).)))).((((((((....)))))(((((....................((((((.....))))))(....)...)))))...)))....... ( -47.60) >Banth.0 291724 120 + 5227293/2200-2320 CCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGA ((((....((((((((....)))))))).)))).((((((((....)))))(((((....................((((((.....))))))(....)...)))))...)))....... ( -47.60) >consensus CCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCACCGGUGGGAUACCACCCUGACUGUAUUGAAAUUCUAACCCACGGCCCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGA ((((....((((((((....)))))))).))))....(((((....)))))(.((((((.((((............((((((.....))))))(....)....))))....)))))).). (-47.80 = -45.42 + -2.37)

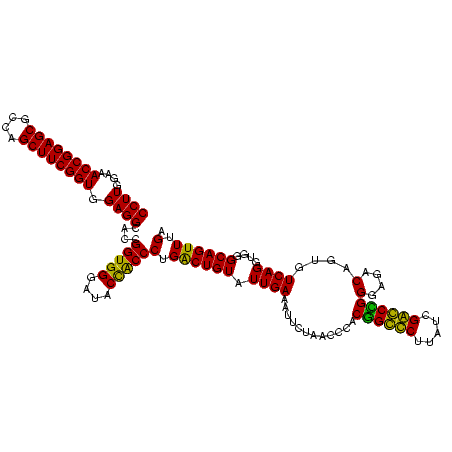
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -52.20 |
| Consensus MFE | -49.38 |
| Energy contribution | -47.50 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.43 |
| SVM RNA-class probability | 0.993869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2240-2360 GGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCA (((..((..((((((((((.((((((((((...((.((((((.....))))))))))).)).)))))....))))))......))))..))(((((((...........)))))))))). ( -50.90) >Bsubt.0 162555 118 + 4214630/2240-2360 GGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUUUCAGGUGGGCAGUUUGACUGGGG-CGGUCGCCUCCUAAA-GGUAACGGAGGCGCCCA (((......))).(((......((((.(((..((((((((((.....))))..(....).......))))))(((.....))))))-.))))((((((....-......))))))))).. ( -50.70) >Bcere.0 332443 119 + 5224283/2240-2360 GGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGG-CGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCA (((..((((((((((((((.((((....((......((((((.....))))))(....)))..))))....)))))).....))))-))))(((((((...........)))))))))). ( -53.60) >Banth.0 291724 119 + 5227293/2240-2360 GGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGG-CGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCA (((..((((((((((((((.((((....((......((((((.....))))))(....)))..))))....)))))).....))))-))))(((((((...........)))))))))). ( -53.60) >consensus GGGAUACCACCCUGACUGUAUUGAAAUUCUAACCCACGGCCCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGG_CGGUCGCCUCCUAAAGGGUAACGGAGGCGCCCA ((....))..((((((....................((((((.....))))))(....)...))))))(((((.....(((((....)))))((((((...........))))))))))) (-49.38 = -47.50 + -1.88)

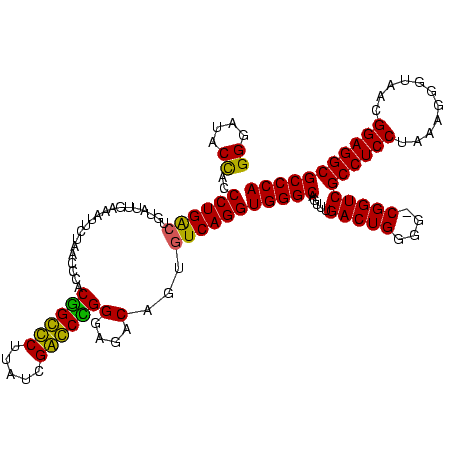
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.38 |
| Mean single sequence MFE | -49.62 |
| Consensus MFE | -45.50 |
| Energy contribution | -43.88 |
| Covariance contribution | -1.62 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.92 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997440 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2240-2360 UGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCC .((((((((((...........))))))....))))........(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))..... ( -49.30) >Bsubt.0 162555 118 + 4214630/2240-2360 UGGGCGCCUCCGUUACC-UUUAGGAGGCGACCG-CCCCAGUCAAACUGCCCACCUGAAACUGUCUCCCCGCCCGAUAAGGGCGGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCC (((((((((((......-....)))))))....-.)))).....(((((((........(((.(((.((((((.....)))))).))).)))..(((.......))).)))))))..... ( -50.80) >Bcere.0 332443 119 + 5224283/2240-2360 UGGGCGCCUCCGUUACACUUUAGGAGGCGACCG-CCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCC .((((((((((...........)))))))((((-(((..((((...........)))).(((.((..((((((.....))))))..)).)))................)))))))..))) ( -49.20) >Banth.0 291724 119 + 5227293/2240-2360 UGGGCGCCUCCGUUACACUUUAGGAGGCGACCG-CCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCC .((((((((((...........)))))))((((-(((..((((...........)))).(((.((..((((((.....))))))..)).)))................)))))))..))) ( -49.20) >consensus UGGGCGCCUCCGUUACACUUUAGGAGGCGACCG_CCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGCCCGAUAAGACCCGCAGGUUAGAAGGUCAAUACAGCCAGGGCAGUAUCCC ..(((((((((...........))))))).))............(((((((..(((...(((.(((.((((((.....)))))).))).))).(......).)))...)))))))..... (-45.50 = -43.88 + -1.62)

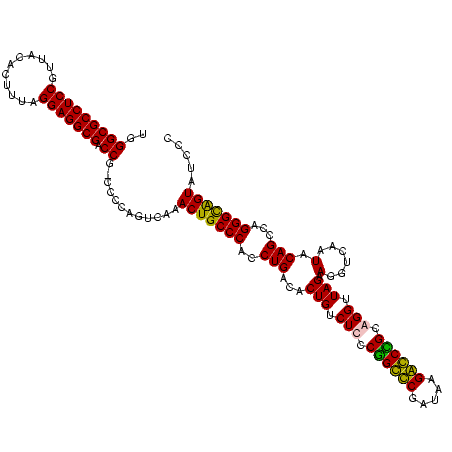
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.65 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -43.65 |
| Energy contribution | -43.15 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.95 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2320-2440 CUGGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGA .((((.((((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))....))))........ ( -46.60) >Bsubt.0 162555 118 + 4214630/2320-2440 CUGGGG-CGGUCGCCUCCUAAA-GGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGA .....(-(((((((((((....-......))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..(((......))).. ( -46.20) >Bcere.0 332443 119 + 5224283/2320-2440 CUGGGG-CGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGA .....(-(((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..(((......))).. ( -45.60) >Banth.0 291724 119 + 5227293/2320-2440 CUGGGG-CGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGA .....(-(((((((((((...........))))))......(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..(((......))).. ( -45.60) >consensus CUGGGG_CGGUCGCCUCCUAAAGGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGA ........((((((((((...........))))))((....(((((((((..(((((((.....))))))).....((((....)))).)))))))))....))..)))).......... (-43.65 = -43.15 + -0.50)

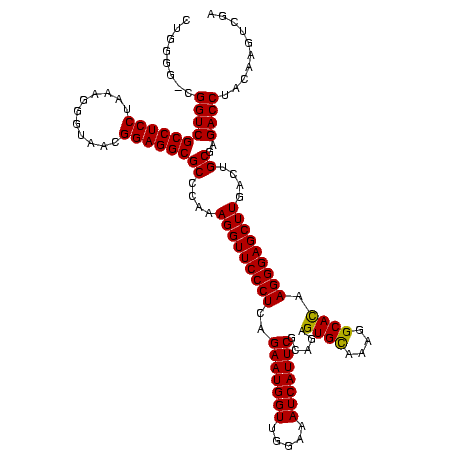
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -42.95 |
| Consensus MFE | -43.45 |
| Energy contribution | -42.95 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.12 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2360-2480 AAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGC .(((((((((..(((((((.....))))))).....((((....)))).)))))))))....(((..(((.((..(((((((...(((....)))..))))...)))...)))))..))) ( -43.20) >Bsubt.0 162555 120 + 4214630/2360-2480 AAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGC .(((((((((..(((((((.....))))))).....((((....)))).)))))))))....(((..(((.((..(((((((...(((....)))..))))...)))...)))))..))) ( -43.20) >Bcere.0 332443 120 + 5224283/2360-2480 AAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGC .(((((((((..(((((((.....))))))).....((((....)))).)))))))))....(((..(((.((..(((((((...(((....)))..))))...)))...)))))..))) ( -42.70) >Banth.0 291724 120 + 5227293/2360-2480 AAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGC .(((((((((..(((((((.....))))))).....((((....)))).)))))))))....(((..(((.((..(((((((...(((....)))..))))...)))...)))))..))) ( -42.70) >consensus AAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGC .(((((((((..(((((((.....))))))).....((((....)))).)))))))))....(((..(((.((..(((((((...(((....)))..))))...)))...)))))..))) (-43.45 = -42.95 + -0.50)

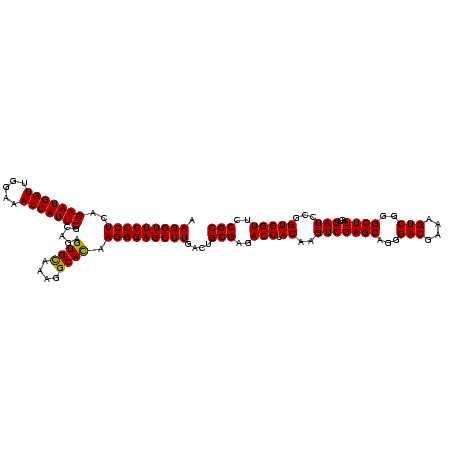
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -47.80 |
| Consensus MFE | -47.80 |
| Energy contribution | -47.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 1.00 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.531542 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2520-2640 GAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAA ........(((.((((((((...((.......))...))))))))..)))....(((((......))))).((((((((((((....))))))).)))))(((((.....)))))..... ( -47.80) >Bsubt.0 162555 120 + 4214630/2520-2640 GAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAA ........(((.((((((((...((.......))...))))))))..)))....(((((......))))).((((((((((((....))))))).)))))(((((.....)))))..... ( -47.80) >Bcere.0 332443 120 + 5224283/2520-2640 GAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAA ........(((.((((((((...((.......))...))))))))..)))....(((((......))))).((((((((((((....))))))).)))))(((((.....)))))..... ( -47.80) >Banth.0 291724 120 + 5227293/2520-2640 GAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAA ........(((.((((((((...((.......))...))))))))..)))....(((((......))))).((((((((((((....))))))).)))))(((((.....)))))..... ( -47.80) >consensus GAUAACAGGCUUAUCUCCCCCAAGAGUCCACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAA ........(((.((((((((...((.......))...))))))))..)))....(((((......))))).((((((((((((....))))))).)))))(((((.....)))))..... (-47.80 = -47.80 + 0.00)

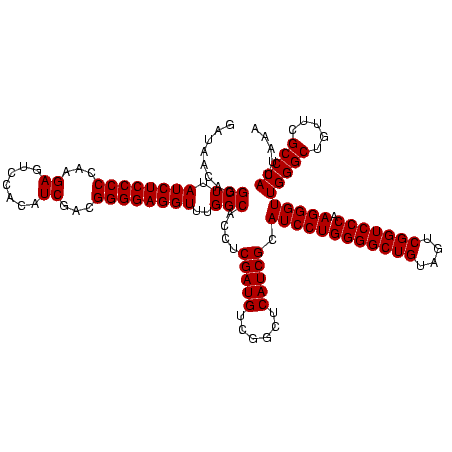
| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -56.60 |
| Consensus MFE | -53.60 |
| Energy contribution | -56.60 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.848832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2560-2680 GAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUC (((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).)))) ( -56.60) >Bsubt.0 162555 120 + 4214630/2560-2680 GAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUC (((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).)))) ( -56.60) >Bcere.0 332443 120 + 5224283/2560-2680 GAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUC (((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).)))) ( -56.60) >Banth.0 291724 120 + 5227293/2560-2680 GAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUC (((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).)))) ( -56.60) >consensus GAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUC (((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..)).(((((((....))).)))) (-53.60 = -56.60 + 3.00)

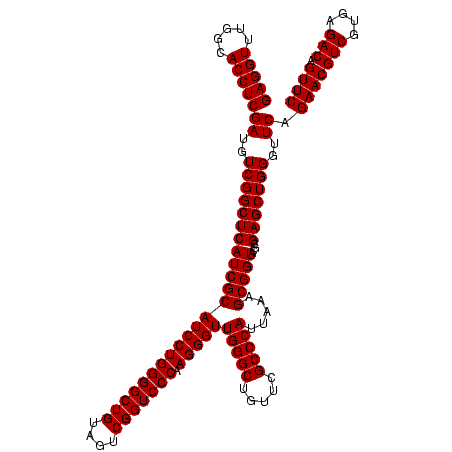

| Location | 159,962 – 160,082 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.84 |
| Mean single sequence MFE | -30.65 |
| Consensus MFE | -30.55 |
| Energy contribution | -29.05 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.65 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.872595 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 120 + 4222334/2800-2920 CCACCUCAGACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUAUAGGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCGUCCGCAC ...((((((.........(((((.......)))))..(((((.((((.....))))))))).........))))))(((((.....)).(((.(((.....))).))).)))........ ( -29.80) >Bsubt.0 162555 119 + 4214630/2800-2920 CCACCUCAGACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUGC-GGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCGUCCGCAC ..................(((((.......))))).............(((-(((.(((((...(((.....)))(..(((.((.......)).)))..)........))))))))))). ( -34.40) >Bcere.0 332443 119 + 5224283/2800-2920 CCACCUCGAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCUUCCGCAC ...((((((.........(((((.......)))))..(((((.((((....-))))))))).........))))))(((((.....)).(((.(((.....))).))).)))........ ( -29.20) >Banth.0 291724 119 + 5227293/2800-2920 CCACCUCGAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCUUCCGCAC ...((((((.........(((((.......)))))..(((((.((((....-))))))))).........))))))(((((.....)).(((.(((.....))).))).)))........ ( -29.20) >consensus CCACCUCAAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUACCUUAC_GCAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCGUCCGCAC ...((((((.........(((((.......)))))..(((((.((((.....))))))))).........))))))(((((.....)).(((.(((.....))).))).)))........ (-30.55 = -29.05 + -1.50)

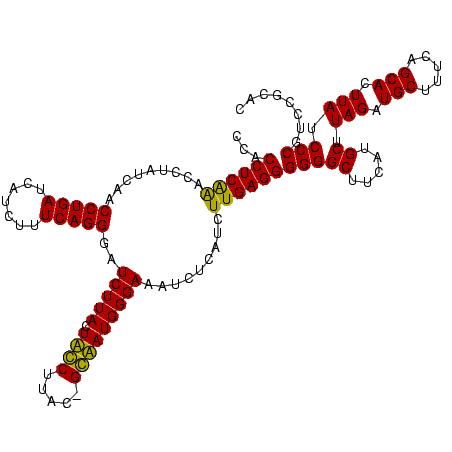
| Location | 159,962 – 160,079 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.42 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -26.95 |
| Energy contribution | -26.45 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.787948 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 159962 117 + 4222334/2880-3000 AGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACCUAAAUAUUUGUGAAUCGUCU---GUCCG ........((.(((((((((((((((((((((((.(((....(((((....)))))...)))..))))).)).))))))).(((......))).........)).)))))))---.)).. ( -33.90) >Bsubt.0 162555 117 + 4214630/2880-3000 AGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACC---AUAUUUUUGAAUGAUGUCACAUCUG ...........(((((....((((((((((((((.(((....(((((....)))))...)))..))))).)).)))))))...((((((..(---(......))..))).))).))))). ( -31.40) >Bcere.0 332443 111 + 5224283/2880-3000 AGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC---------AUAUAAUAUGAAGCAAAUG .(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))..((......))---------................... ( -27.60) >Banth.0 291724 111 + 5227293/2880-3000 AGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC---------AUAUAAUAUGUAGCAAAUG .(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))..((......))---------................... ( -27.60) >consensus AGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCCGAGGUGGAAGCAUGGCGACACAUGGAGCUGACAAAUACUAAUAGAUCGAGGACUUAACC_________AUAAAAUAUGUAGCAAAUG .(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))........................................ (-26.95 = -26.45 + -0.50)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:23:49 2006