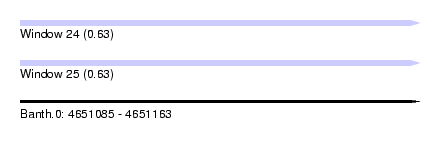
| Sequence ID | Banth.0 |
|---|---|
| Location | 4,651,085 – 4,651,163 |
| Length | 78 |
| Max. P | 0.633421 |

| Location | 4,651,085 – 4,651,163 |
|---|---|
| Length | 78 |
| Sequences | 4 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 96.58 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -31.16 |
| Energy contribution | -31.23 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4651085 78 + 5227293 AAGUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAGGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCAU ..((((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))) ( -31.60) >Bcere.0 4650063 78 + 5224283 AAGUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAGGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCAU ..((((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))) ( -31.60) >Bhalo.0 2949474 77 + 4202352 G-GUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCAU .-((((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))) ( -32.30) >Bclau.0 2627848 78 + 4303871 AAGUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCGA ...(((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).))). ( -30.80) >consensus AAGUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCAU ..((((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))) (-31.16 = -31.23 + 0.06)

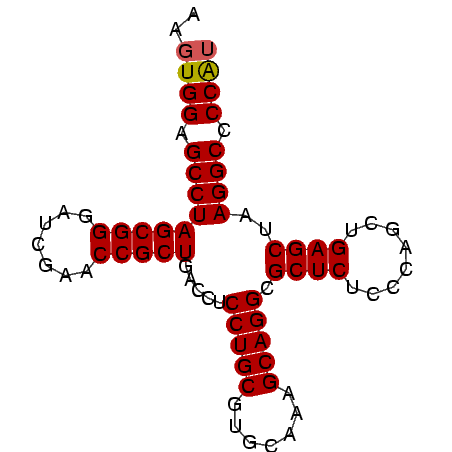
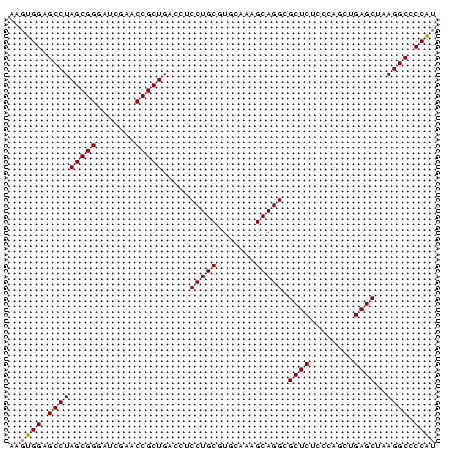
| Location | 4,651,085 – 4,651,163 |
|---|---|
| Length | 78 |
| Sequences | 4 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 96.58 |
| Mean single sequence MFE | -31.57 |
| Consensus MFE | -31.16 |
| Energy contribution | -31.23 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633421 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 4651085 78 + 5227293/0-78 AAGUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAGGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCAU ..((((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))) ( -31.60) >Bcere.0 4650063 78 + 5224283/0-78 AAGUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAGGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCAU ..((((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))) ( -31.60) >Bhalo.0 2949474 77 + 4202352/0-78 G-GUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCAU .-((((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))) ( -32.30) >Bclau.0 2627848 78 + 4303871/0-78 AAGUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCGA ...(((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).))). ( -30.80) >consensus AAGUGGAGCCUAGCGGGAUCGAACCGCUGACCUCCUGCGUGCAAAGCAGGCGCUCUCCCAGCUGAGCUAAGGCCCCAU ..((((.(((((((((.......))))).....(((((.......))))).((((........))))..)))).)))) (-31.16 = -31.23 + 0.06)

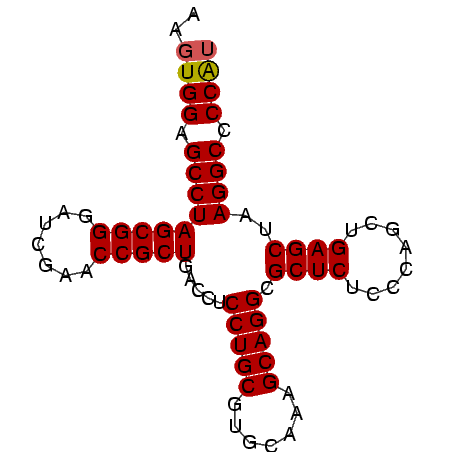
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:15:11 2006