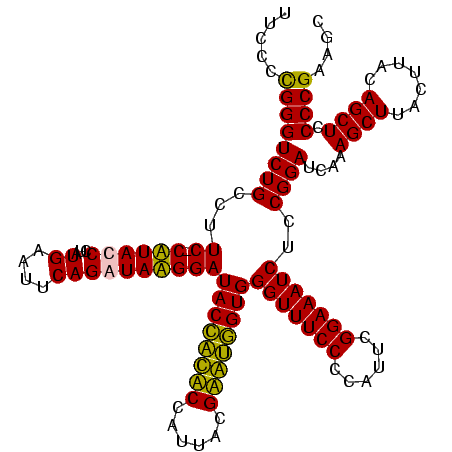
| Sequence ID | Bcere.0 |
|---|---|
| Location | 84,077 – 84,197 |
| Length | 120 |
| Max. P | 0.999265 |

| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -42.90 |
| Consensus MFE | -44.35 |
| Energy contribution | -41.85 |
| Covariance contribution | -2.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.38 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.75 |
| SVM RNA-class probability | 0.996793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
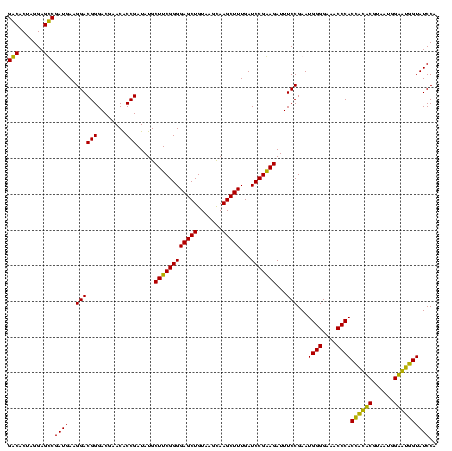
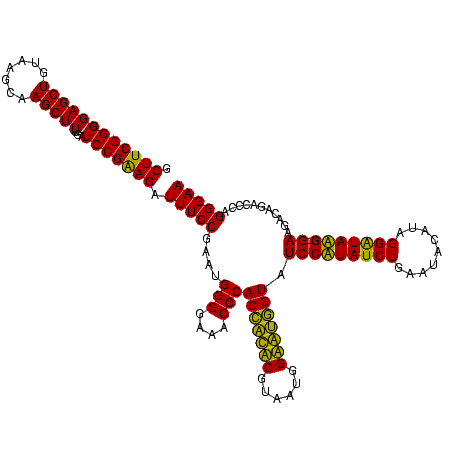
>Bcere.0 84077 120 + 5224283/80-200 GACACUAGGAGUCGAUGAAGGACGGGACUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCU ......((((.(((.....((.((.......))))((....((((((((((((.......)))))...)))))))...)))))..(((....)))(((((((......))))))).)))) ( -40.60) >Banth.0 84076 120 + 5227293/80-200 GACACUAGGAGUCGAUGAAGGACGGGACUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCU ......((((.(((.....((.((.......))))((....((((((((((((.......)))))...)))))))...)))))..(((....)))(((((((......))))))).)))) ( -40.60) >Blich.0 96991 120 + 4222334/80-200 GGCACUAGGAGCCGAUGAAGGACGGGACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCA (((.......)))......((((((((((.....)).....((((((((((((.......)))))...)))))))..)))))...(((....)))(((((((......))))))).))). ( -45.20) >Bsubt.0 92183 120 + 4214630/80-200 GGCACUAGGAGCCGAUGAAGGACGGGACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCA (((.......)))......((((((((((.....)).....((((((((((((.......)))))...)))))))..)))))...(((....)))(((((((......))))))).))). ( -45.20) >consensus GACACUAGGAGCCGAUGAAGGACGGGACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCACACGUAAUGGAAUGGUAUCCA (((.......)))((((..((((((........))).....((((((((((((.......)))))...)))))))...)))...((((....))))((((((......)))))))))).. (-44.35 = -41.85 + -2.50)


| Location | 84,077 – 84,196 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -45.35 |
| Consensus MFE | -44.85 |
| Energy contribution | -42.10 |
| Covariance contribution | -2.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -4.04 |
| Structure conservation index | 0.99 |
| SVM decision value | 3.15 |
| SVM RNA-class probability | 0.998598 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
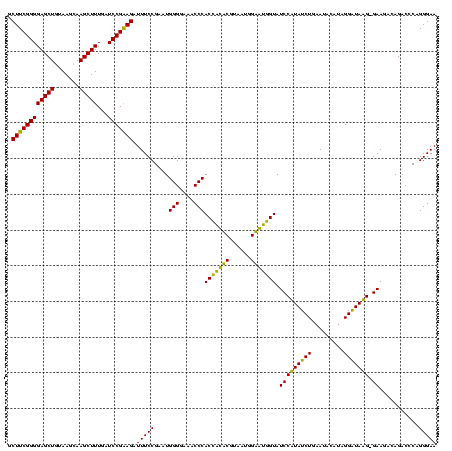
>Bcere.0 84077 119 + 5224283/120-240 GCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACAGACCCAGGGAA .((((((((((((.......)))))...))))))).(((((....(((....)))(((((((......))))))).(((((((((.........)))))))-))...........))))) ( -44.80) >Banth.0 84076 119 + 5227293/120-240 GCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACAGACCCAGGGAA .((((((((((((.......)))))...))))))).(((((....(((....)))(((((((......))))))).(((((((((.........)))))))-))...........))))) ( -44.80) >Blich.0 96991 120 + 4222334/120-240 GCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAA .(((((((..((.....))..(((((....(((......)))...(((....)))(((((((......)))))))...(((((((.........)))))))..)))))...))))))).. ( -45.90) >Bsubt.0 92183 120 + 4214630/120-240 GCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAA .(((((((..((.....))..(((((....(((......)))...(((....)))(((((((......)))))))...(((((((.........)))))))..)))))...))))))).. ( -45.90) >consensus GCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCACACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAAG_GAAGACAGACCCAGGGAA .((((((((((((.......)))))...))))))).(((((....(((....)))(((((((......))))))).(((((((((.........))))))).))...........))))) (-44.85 = -42.10 + -2.75)

| Location | 84,077 – 84,196 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.10 |
| Mean single sequence MFE | -33.25 |
| Consensus MFE | -31.70 |
| Energy contribution | -29.70 |
| Covariance contribution | -2.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.990709 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 119 + 5224283/120-240 UUCCCUGGGUCUGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGC ((((.((((((((.....-..(((.....)))..)))))..(((.(((.((((((.....))))))))).))))))...))))..((((((.....((((.......))))..)))))). ( -31.90) >Banth.0 84076 119 + 5227293/120-240 UUCCCUGGGUCUGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGC ((((.((((((((.....-..(((.....)))..)))))..(((.(((.((((((.....))))))))).))))))...))))..((((((.....((((.......))))..)))))). ( -31.90) >Blich.0 96991 120 + 4222334/120-240 UUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGC .....((((...((....(((((...))))).....(((.((((((((((((......))))))))(((((((......))))))))))).)))..............))..)))).... ( -34.60) >Bsubt.0 92183 120 + 4214630/120-240 UUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGC .....((((...((....(((((...))))).....(((.((((((((((((......))))))))(((((((......))))))))))).)))..............))..)))).... ( -34.60) >consensus UUCCCCGGGUCUGCCUUC_CAUACCCUAUGAAUUCAGAUAAGGAUACCACACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUACUUACAGCUCCCCGAAGC .....((((((((...((.((((((...((....))))))))))((((((((......))))))))(((((((......)))))))..))))....((((.......)))).)))).... (-31.70 = -29.70 + -2.00)

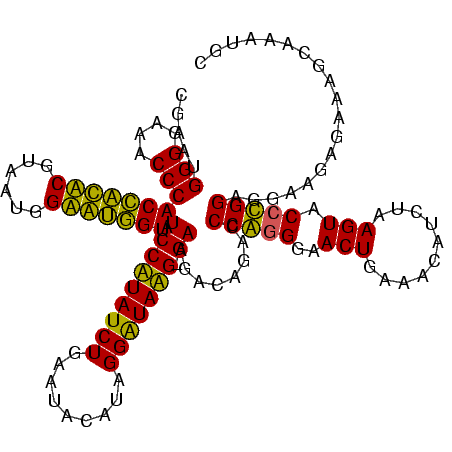
| Location | 84,077 – 84,196 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -36.40 |
| Energy contribution | -33.65 |
| Covariance contribution | -2.75 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.96 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998555 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 119 + 5224283/160-280 CGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACGUGGAGGAAGAGAAAGCAAAUGC ....((((....))))((.((((((..(((.((...(((((((((.........)))))))-))...)).)))(((....)))...........))))))..))................ ( -35.30) >Banth.0 84076 119 + 5227293/160-280 CGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGAAGAGAAAGCAAAUGC .....(((....)))(((((((......))))))).(((((((((.........)))))))-))........(((((..(((..........))).)))))................... ( -36.60) >Blich.0 96991 120 + 4222334/160-280 CGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGC .....(((....)))(((((((......))))))).(((...((((..(((.((.....)).)))..)))).(((((..(((..........))).))))).)))............... ( -39.90) >Bsubt.0 92183 120 + 4214630/160-280 CGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGC .....(((....)))(((((((......))))))).(((...((((..(((.((.....)).)))..)))).(((((..(((..........))).))))).)))............... ( -39.90) >consensus CGAAUGGGGAAACCCACCACACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAAG_GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCCGGAGGAAGAGAAAGCAAAUGC .....(((....)))(((((((......))))))).(((((((((.........))))))).))........(((((..(((..........))).)))))................... (-36.40 = -33.65 + -2.75)


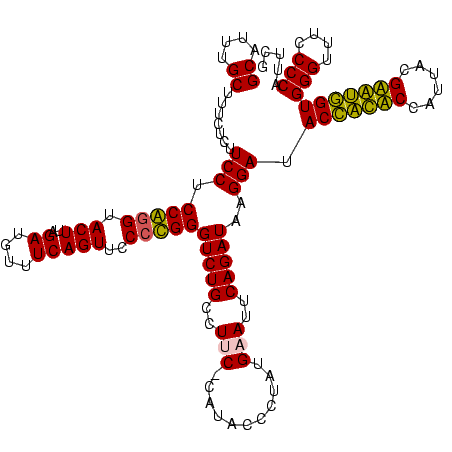
| Location | 84,077 – 84,196 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -29.02 |
| Consensus MFE | -29.30 |
| Energy contribution | -27.80 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.61 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.927850 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 119 + 5224283/160-280 GCAUUUGCUUUCUCUUCCUCCACGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCG ((....))..............(((((..........(((....)))(((.((...((-((((..((........)).))))))...)).))).....)))))(((((....)))))... ( -22.20) >Banth.0 84076 119 + 5227293/160-280 GCAUUUGCUUUCUCUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCG ((....)).......(((((((((.(((..((....)))))..)))))(((((.....-..(((.....)))..))))).)))).(((((((......)))))))(((....)))..... ( -27.70) >Blich.0 96991 120 + 4222334/160-280 GCAUUUGCUUUCUCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCG ((....)).......(((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..))).(((((((......)))))))(((....)))..... ( -33.10) >Bsubt.0 92183 120 + 4214630/160-280 GCAUUUGCUUUCUCUUCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCG ((....)).......(((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..))).(((((((......)))))))(((....)))..... ( -33.10) >consensus GCAUUUGCUUUCUCUUCCUCCAGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUC_CAUACCCUAUGAAUUCAGAUAAGGAUACCACACCAUUACGAAUGGUGGGUUUCCCCAUUCG ((....)).......(((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..))).(((((((......)))))))(((....)))..... (-29.30 = -27.80 + -1.50)

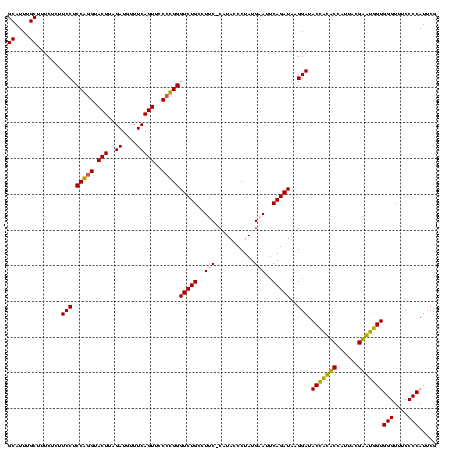
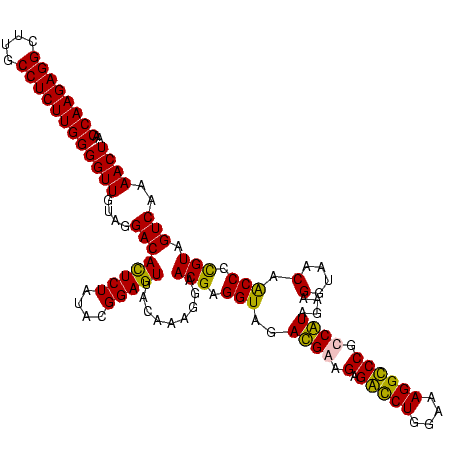
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -40.50 |
| Consensus MFE | -37.60 |
| Energy contribution | -37.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/320-440 AACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCCCGUAGUCGAAACU ....((((((....))))))((((((((...((.((((((.....(((.......)))......((((....(((((....))))))))))))))).)).))))))))............ ( -41.20) >Banth.0 84076 120 + 5227293/320-440 AACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCCCGUAGUCGAAACU ....((((((....))))))((((((((...((.((((((.....(((.......)))......((((....(((((....))))))))))))))).)).))))))))............ ( -41.20) >Blich.0 96991 120 + 4222334/320-440 AACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUGUACGGAGUUGCAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGCCCUGUAGUCAAAACU ..((((((((....))))))))((((....((((((((....))))).........(((.(((.(.....(.(((((....))))))(((......)))..).))).))).)))..)))) ( -39.80) >Bsubt.0 92183 120 + 4214630/320-440 AACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUGUACGGAGUUACAAAGGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGCCCUGUAGUCAAAACU ..((((((((....))))))))((((....((((((((....))))).........(((.(((.(.....(.(((((....))))))(((......)))..).))).))).)))..)))) ( -39.80) >consensus AACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGAGACCUGGAAAGGCCCGCCAUAGAAGGUAACAACCCCGUAGUCAAAACU ..((((((((....))))))))((((....((((((((....))))).........(((.(((..((((.(.(((((....)))))).))))....(....).))).))).)))..)))) (-37.60 = -37.60 + 0.00)

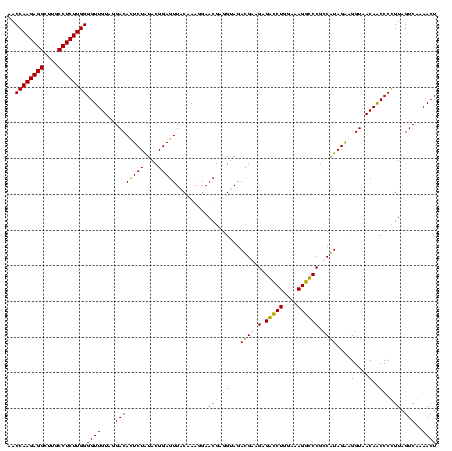
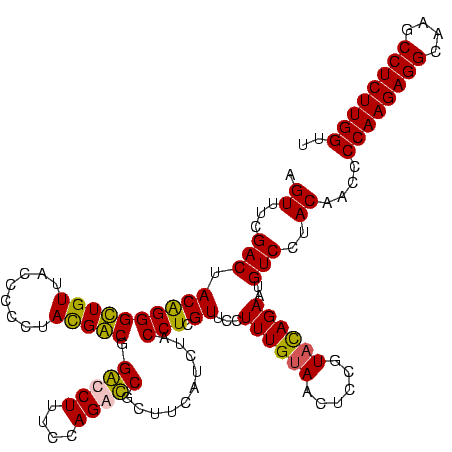
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.39 |
| Mean single sequence MFE | -33.02 |
| Consensus MFE | -28.85 |
| Energy contribution | -27.85 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/320-440 AGUUUCGACUACGGGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUACAACCCCAAGAGGCAAGCCUCUUGGUU ...(((...(((((((((((........))(((((((((....)))))....))))................)))))))))..)))............((((((((....)))))))).. ( -35.80) >Banth.0 84076 120 + 5227293/320-440 AGUUUCGACUACGGGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUACAACCCCAAGAGGCAAGCCUCUUGGUU ...(((...(((((((((((........))(((((((((....)))))....))))................)))))))))..)))............((((((((....)))))))).. ( -35.80) >Blich.0 96991 120 + 4222334/320-440 AGUUUUGACUACAGGGCUGUUACCUCCUAUGGCGGGCCUUUCCAGACCUCUUCAUCUACCUCGUUCUUUUGCAACUCCGUACAGAGUGUCCUACAACCCCAAGAGGCAAGCCUCUUGGUU ....(((.....(((.((((((.......)))))).))).....(((((((.....(((...((((....).)))...))).)))).)))...)))..((((((((....)))))))).. ( -29.10) >Bsubt.0 92183 120 + 4214630/320-440 AGUUUUGACUACAGGGCUGUUACCUCCUAUGGCGGGCCUUUCCAGACCUCUUCAUCUACCUCGUUCCUUUGUAACUCCGUACAGAGUGUCCUACAACCCCAAGAGGCAAGCCUCUUGGUU .((...(((...(((.((((((.......)))))).)))...........................(((((((......))))))).)))..))....((((((((....)))))))).. ( -31.40) >consensus AGUUUCGACUACAGGGCUGUUACCCCCUACGACGGACCUUUCCAGACCGCUUCAUCUACCUCGUUCCUUUGUAACUCCGUACAGAAUGUCCUACAACCCCAAGAGGCAAGCCUCUUGGUU .((...(((.((((((((((........))))).(((((....)))))..........))).))...((((((......))))))..)))..))....((((((((....)))))))).. (-28.85 = -27.85 + -1.00)


| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.03 |
| Mean single sequence MFE | -39.55 |
| Consensus MFE | -39.51 |
| Energy contribution | -39.07 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/560-680 GGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUU ((((..(.(....).).((((....))))...........))))....(((.((..(((((((..((((..(((((....)))))..))))......))))))).....)))))...... ( -40.00) >Banth.0 84076 120 + 5227293/560-680 GGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAAUGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUU ((((..(.(....).).((((....))))...........))))....(((.((..(((((((..((((..(((((....)))))..))))......))))))).....)))))...... ( -37.40) >Blich.0 96991 120 + 4222334/560-680 GGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUU ((((..(.(....).).((((....))))...........))))....(((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....)))))...... ( -40.40) >Bsubt.0 92183 120 + 4214630/560-680 GGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUU ((((..(.(....).).((((....))))...........))))....(((.((..(((((((((((((..(((((....)))))..)))).)))...)))))).....)))))...... ( -40.40) >consensus GGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUU ((((..........((.((((....)))).))........))))....(((.((..(((((((..((((..(((((....)))))..))))......))))))).....)))))...... (-39.51 = -39.07 + -0.44)

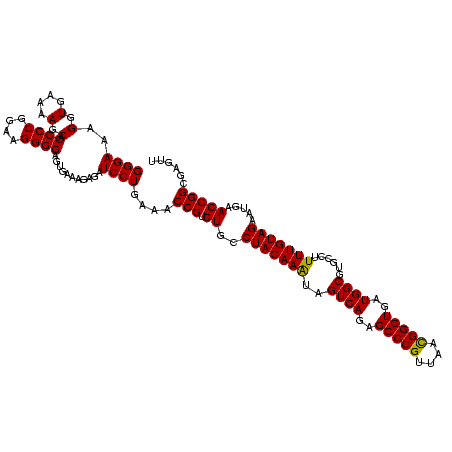
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -47.75 |
| Consensus MFE | -46.35 |
| Energy contribution | -46.35 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.51 |
| SVM RNA-class probability | 0.959851 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/760-880 GUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUU ....(((.(....).)))...((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))).......... ( -45.90) >Banth.0 84076 120 + 5227293/760-880 GUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUU ....(((.(....).)))...((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))).......... ( -45.90) >Blich.0 96991 120 + 4222334/760-880 GUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUG ....(((((....).......((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))))))))))...... ( -49.60) >Bsubt.0 92183 120 + 4214630/760-880 GUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUG ....(((((....).......((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..))))))))))))...... ( -49.60) >consensus GUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUG ....(((.(....).)))...((((((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))))))).......... (-46.35 = -46.35 + 0.00)

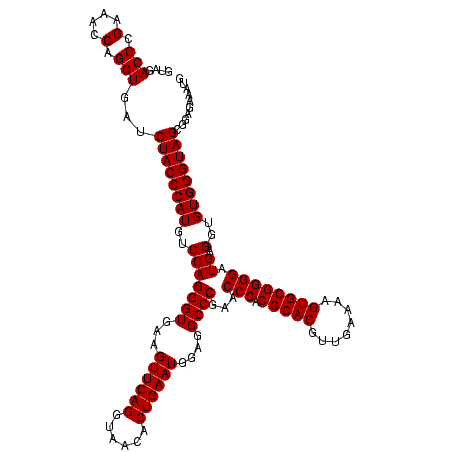
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -39.35 |
| Consensus MFE | -39.35 |
| Energy contribution | -39.35 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.43 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/760-880 AAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUAC ..........(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))).... ( -39.30) >Banth.0 84076 120 + 5227293/760-880 AAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUAC ..........(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))).... ( -39.30) >Blich.0 96991 120 + 4222334/760-880 CAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUAC ..........(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))).... ( -39.40) >Bsubt.0 92183 120 + 4214630/760-880 CAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUAC ..........(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))).... ( -39.40) >consensus AAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGGGUAGAUCACCUGGUUUCGGGUCUAC ..........(((((((.........(((((((........))))).))(((((((......(((((......)))))......))))))).)))))))...(((((....))))).... (-39.35 = -39.35 + 0.00)

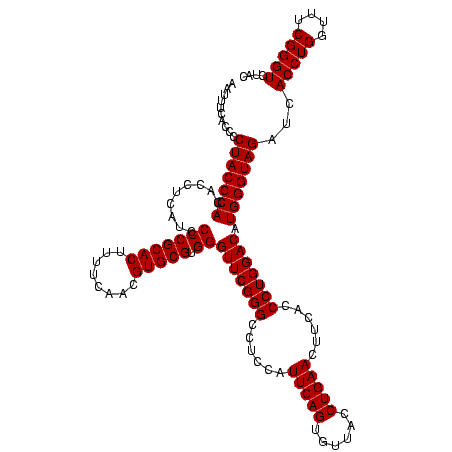
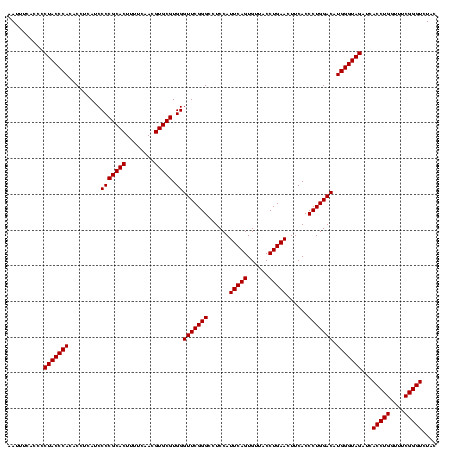
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -33.25 |
| Energy contribution | -33.00 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/800-920 GCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACU ........(((((((........)))))((((((((..(((.....)))................((((((((........))))).))).)))))))))).(((((......))))).. ( -37.40) >Banth.0 84076 120 + 5227293/800-920 GCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACU ........(((((((........)))))((((((((..(((.....)))................((((((((........))))).))).)))))))))).(((((......))))).. ( -37.40) >Blich.0 96991 120 + 4222334/800-920 GCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACU ........((((.(((((...........))))).............(((..((((((...........((((........))))))))))..))).)))).(((((......))))).. ( -33.30) >Bsubt.0 92183 120 + 4214630/800-920 GCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACU ........((((.(((((...........))))).............(((..((((((...........((((........))))))))))..))).)))).(((((......))))).. ( -33.30) >consensus GCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACU ........(((((..........)))))((((((((..((.......))................((((((((........))))).))).))))))))...(((((......))))).. (-33.25 = -33.00 + -0.25)

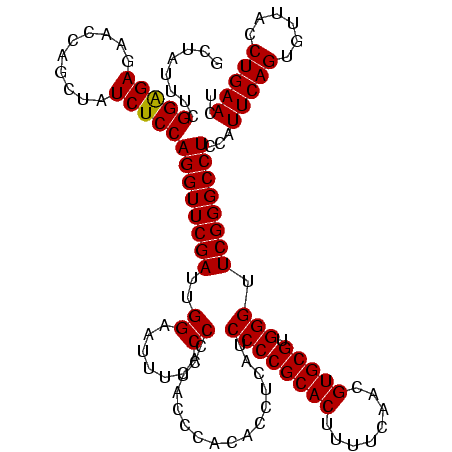
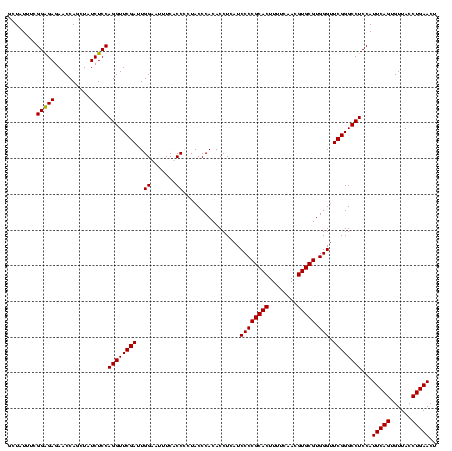
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.13 |
| Mean single sequence MFE | -38.30 |
| Consensus MFE | -37.20 |
| Energy contribution | -36.20 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.780100 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/920-1040 UUUAGGGCUAGCCUUAAGUGUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCUCAUCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCA .....((((...((((((.(...(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))))))). ( -34.60) >Banth.0 84076 120 + 5227293/920-1040 UUUAGGGCUAGCCUUAAGUGUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGUCCUCAUCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCA .....((((...((((((.(...(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))))))). ( -34.60) >Blich.0 96991 119 + 4222334/920-1040 UUUAGGGCUAGCCUCAAG-GUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGCCCCUACCGGGUUACCGAAUUCAGUCAAACUCCGAAUGCCAAAGACUUAUCCUUGGGAGUCA .....((((...((((((-(...(((((((...(((((((...((((((((...(((((((.....))))).))...))))))))..)))....)))).)))))))..))))))))))). ( -42.40) >Bsubt.0 92183 119 + 4214630/920-1040 UUUAGGGCUAGCCUCAAG-GUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGCCCCUACCGGGUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUGGGAGUCA .....((((...((((((-(...(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))))))). ( -41.60) >consensus UUUAGGGCUAGCCUCAAG_GUAAGAGUCUUGGAGGUAGAGCACUGAUUGGACUAGGGGCCCCCACCGGAUUACCGAAUUCAGUCAAACUCCGAAUGCCAAUGACUUAUCCUUAGGAGUCA .....((((...((((((.(...(((((((((.....(((...((((((((...(((((((.....))))).))...))))))))..)))......)))).)))))..))))))))))). (-37.20 = -36.20 + -1.00)

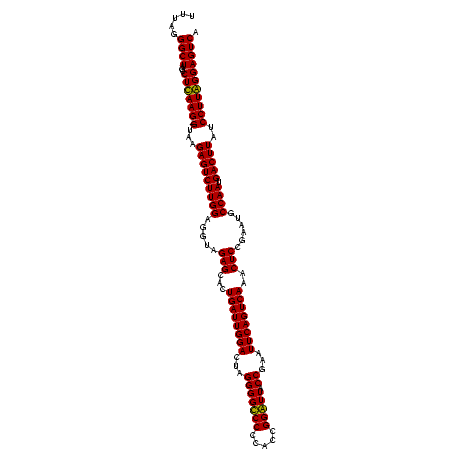
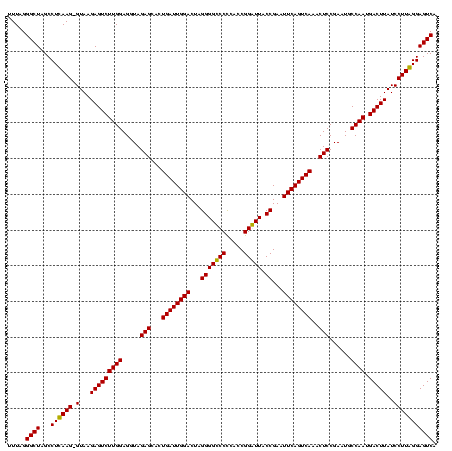
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -37.30 |
| Energy contribution | -36.80 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.99 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.844431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1120-1240 AAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAU ....((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..))))..................... ( -37.10) >Banth.0 84076 120 + 5227293/1120-1240 AAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAU ....((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..))))..................... ( -37.10) >Blich.0 96991 120 + 4222334/1120-1240 AAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAC ....((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..))))..................... ( -37.80) >Bsubt.0 92183 120 + 4214630/1120-1240 AAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAC ....((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..))))..................... ( -37.80) >consensus AAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAC ....((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..))))..................... (-37.30 = -36.80 + -0.50)

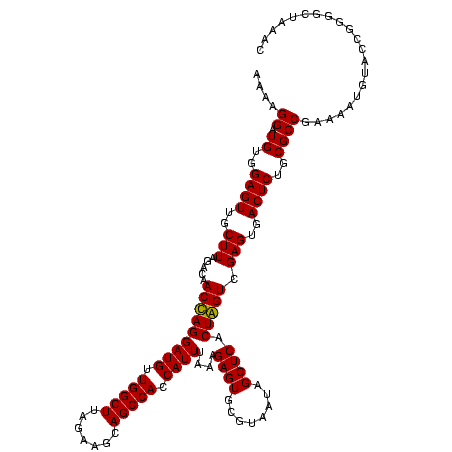
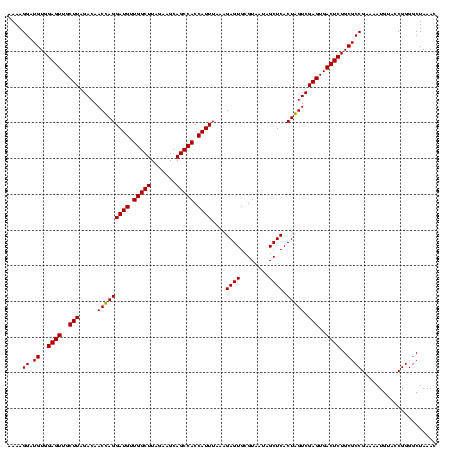
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.76 |
| Mean single sequence MFE | -37.03 |
| Consensus MFE | -33.49 |
| Energy contribution | -33.80 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.817480 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1160-1280 ACUGAUACCAUUCGGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUA ..(((((((....)))))))....((((((((((.(((.......)))))))..(((((..((.((..((((....)))).((((....)))))).))....)))))..))))))..... ( -37.70) >Banth.0 84076 119 + 5227293/1160-1280 ACUGAUACCAUU-GGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACUAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUA ..(((((((...-)))))))....((((((((((.(((.......)))))))..(((((..((.((..((((....)))).((((....)))))).))....)))))..))))))..... ( -36.10) >Blich.0 96991 116 + 4222334/1160-1280 ACUGUUCGCA----AGAACAGUCCGCAGCUUCGGUGAUACGUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUA (((((((...----.)))))))..(((((....((((((.(((((((.((((.......)))).)).(.(((....))))..))))))))))).((((........)))))))))..... ( -39.80) >Bsubt.0 92183 115 + 4214630/1160-1280 ACUGUUCG-A----AGAACAGUCCGCAGCUUCGGUGAUACGUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUA (((((((.-.----.)))))))..(((((....((((((.(((((((.((((.......)))).)).(.(((....))))..))))))))))).((((........)))))))))..... ( -34.50) >consensus ACUGAUACCA____AGAACAAUCCGCAGCUUCGGUGAUACAUUUAGCCCCGGUACAUUUUCGGCGCAGAGUCACUCGACCAGUGAGCUAUUACGCACUCUUUAAAUGGUGGCUGCUUCUA (((((((((....)))))))))..((((((.(.((..........((.((((.......)))).))(((((((((.....)))))((......)).))))....)).).))))))..... (-33.49 = -33.80 + 0.31)

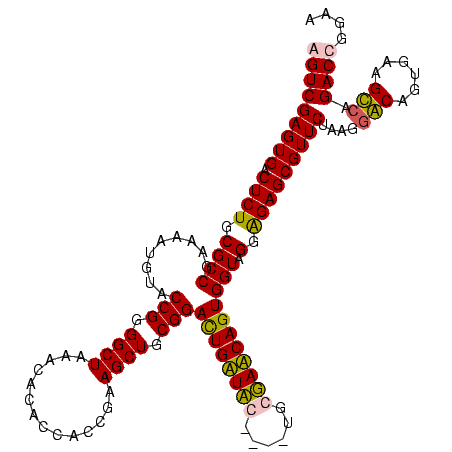
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.10 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -37.06 |
| Energy contribution | -36.32 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1200-1320 AGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACCGAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAA .((((((((.((((.((((.........(((.((((..............)))).)))(((((((((....)))))))))))).).)))))))))....(((......))).)))..... ( -37.64) >Banth.0 84076 119 + 5227293/1200-1320 AGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACC-AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAA .((((((((.((((.((((.........(((.((((..............)))).)))(((((((((-...)))))))))))).).)))))))))....(((......))).)))..... ( -36.44) >Blich.0 96991 116 + 4222334/1200-1320 GGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAUCACCGAAGCUGCGGACUGUUCU----UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAA (((((((((.((((.((((.........(((.((((...((......)).)))).)))(((((((.----...)))))))))).).))))))))).....(((....)))..)))).... ( -41.00) >Bsubt.0 92183 115 + 4214630/1200-1320 GGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACGUAUCACCGAAGCUGCGGACUGUUCU----U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAA (((((((((.((((.((((.........(((.((((...((......)).)))).)))(((((((.----.-.)))))))))).).)))))))))....((((....)))).)))).... ( -42.80) >consensus AGUCGAGUGACUCUGCGCCGAAAAUGUACCGGGGCUAAACACACCACCGAAGCUGCGGACUGAUAC____UGCGAACAGUGGUAGGAGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAA (((((((((.((((.((((.........(((.((((..............)))).)))((((((((......))))))))))).).)))))))))....(((......))).)))).... (-37.06 = -36.32 + -0.75)

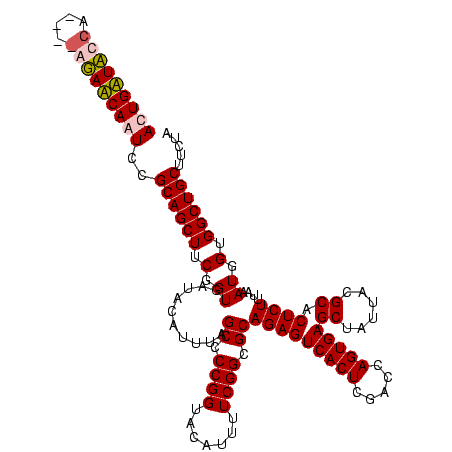
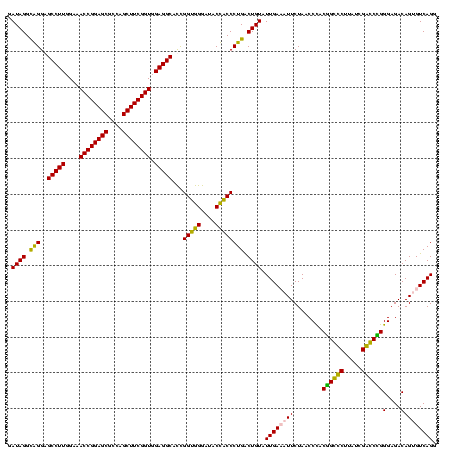
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.24 |
| Mean single sequence MFE | -42.83 |
| Consensus MFE | -43.08 |
| Energy contribution | -41.20 |
| Covariance contribution | -1.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.11 |
| Structure conservation index | 1.01 |
| SVM decision value | 3.47 |
| SVM RNA-class probability | 0.999265 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1240-1360 ACACCACCGAAGCUGCGGAUUGAUACCGAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAG ..(((.(((......)))(((((((((....)))))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))))..... ( -40.20) >Banth.0 84076 119 + 5227293/1240-1360 ACACCACCGAAGCUGCGGAUUGAUACC-AAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAG ..(((.(((......)))(((((((((-...)))))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))))..... ( -39.00) >Blich.0 96991 116 + 4222334/1240-1360 GUAUCACCGAAGCUGCGGACUGUUCU----UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAG (((((.(((......)))(((((((.----...)))))))((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))))))))... ( -46.50) >Bsubt.0 92183 115 + 4214630/1240-1360 GUAUCACCGAAGCUGCGGACUGUUCU----U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAG (((((.(((......)))(((((((.----.-.)))))))((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))))))))... ( -45.60) >consensus ACACCACCGAAGCUGCGGACUGAUAC____UGCGAACAGUGGUAGGAGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAG (((((.(((......)))((((((((......))))))))((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))))))... (-43.08 = -41.20 + -1.88)

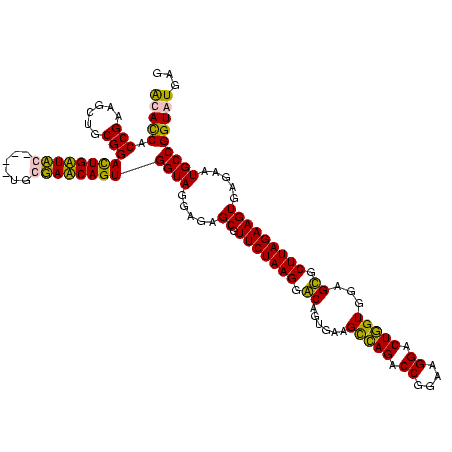
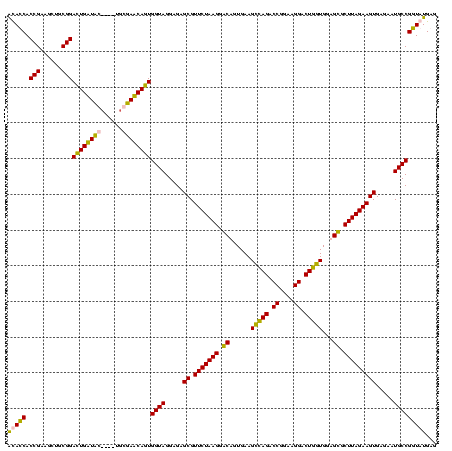
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.24 |
| Mean single sequence MFE | -31.14 |
| Consensus MFE | -27.81 |
| Energy contribution | -27.75 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.91 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995649 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1240-1360 CUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUCGGUAUCAAUCCGCAGCUUCGGUGGUGU ...(((((.((........(((((((.((.....(((.((....)).)))........)).)))))))..............(((((((....)))))))....)).((....))))))) ( -29.52) >Banth.0 84076 119 + 5227293/1240-1360 CUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUU-GGUAUCAAUCCGCAGCUUCGGUGGUGU ...(((((.((........(((((((.((.....(((.((....)).)))........)).)))))))..............(((((((...-)))))))....)).((....))))))) ( -27.92) >Blich.0 96991 116 + 4222334/1240-1360 CUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCGCA----AGAACAGUCCGCAGCUUCGGUGAUAC ....((((((.........(((((((.(((...((((.((....)).))))......))).))))))).(((........(((((((...----.)))))))....))).)))))).... ( -35.90) >Bsubt.0 92183 115 + 4214630/1240-1360 CUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCG-A----AGAACAGUCCGCAGCUUCGGUGAUAC ....((((((.........(((((((.(((...((((.((....)).))))......))).))))))).(((........(((((((.-.----.)))))))....))).)))))).... ( -31.20) >consensus CUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACAGCCCUUAGAACGCUCCCCUACCACUGAUACCA____AGAACAAUCCGCAGCUUCGGUGAUAC ....((((((.........(((((((.((....((((.((....)).)))).......)).))))))).(((........(((((((((....)))))))))....))).)))))).... (-27.81 = -27.75 + -0.06)

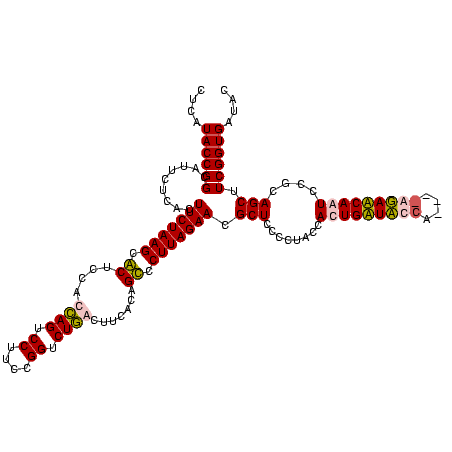
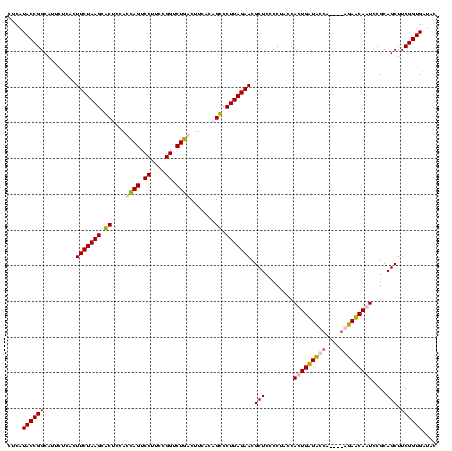
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -44.20 |
| Consensus MFE | -44.92 |
| Energy contribution | -43.30 |
| Covariance contribution | -1.62 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.17 |
| Structure conservation index | 1.02 |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.997177 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1280-1400 GGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAAUGCCUA ((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))(((((......((...((((((.......))))))...)).))))).. ( -39.30) >Banth.0 84076 120 + 5227293/1280-1400 GGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCGUCCACCGAAUGCCUA ((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))(((((......((...((((((.......))))))...)).))))).. ( -39.30) >Blich.0 96991 120 + 4222334/1280-1400 GGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGGGGUGAGAAUCCCCUCCACCGAAUGCCUA ((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....))))(((((......((...((((((.......))))))...)).))))).. ( -48.80) >Bsubt.0 92183 120 + 4214630/1280-1400 GGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGGGGUGAGAAUCCCCUCCACCGAAUGCCUA ((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....))))(((((......((...((((((.......))))))...)).))))).. ( -49.40) >consensus GGUAGGAGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCCUCCACCGAAUGCCUA ((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))(((((......((...((((((.......))))))...)).))))).. (-44.92 = -43.30 + -1.62)

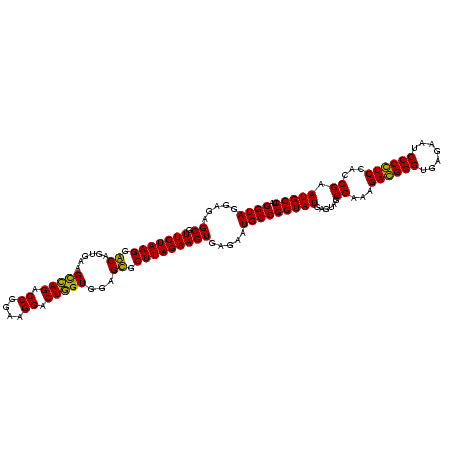
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.28 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -32.08 |
| Energy contribution | -31.20 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.993009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1280-1400 UAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACC ((((....(((((((((((.......))))))....(((.........))).....)))(((((((.((.....(((.((....)).)))........)).)))))))))....)))).. ( -29.02) >Banth.0 84076 120 + 5227293/1280-1400 UAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACC ((((....(((((((((((.......))))))....(((.........))).....)))(((((((.((.....(((.((....)).)))........)).)))))))))....)))).. ( -29.02) >Blich.0 96991 120 + 4222334/1280-1400 UAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUCCUACC ((((....(((((((((((.......))))))....(((.........))).....)))(((((((.(((...((((.((....)).))))......))).)))))))))....)))).. ( -35.40) >Bsubt.0 92183 120 + 4214630/1280-1400 UAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUCCUACC ((((....(((((((((((.......))))))....(((.........))).....)))(((((((.(((...((((.((....)).))))......))).)))))))))....)))).. ( -36.00) >consensus UAGGCAUUCGGUGGACGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACAGCCCUUAGAACGCUCCCCUACC ((((....(((((((((((.......))))))....(((.........))).....)))(((((((.((....((((.((....)).)))).......)).)))))))))....)))).. (-32.08 = -31.20 + -0.88)

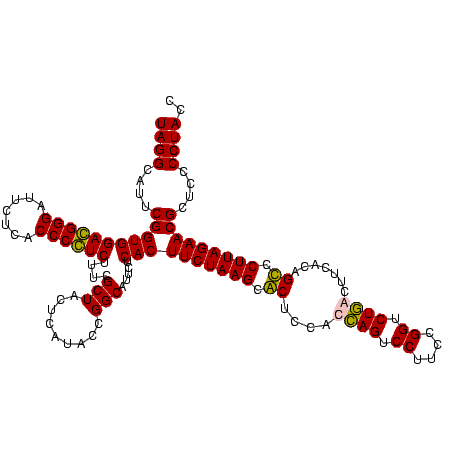
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -42.40 |
| Consensus MFE | -42.50 |
| Energy contribution | -42.00 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1360-1480 UGUUGUCCAUCGCCUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUA ..((((((...(((.((....)).)))....(((((((...........)))))))))))))((((......(....)...))))...(((.(((((((.......))))))).)))... ( -42.40) >Banth.0 84076 120 + 5227293/1360-1480 UGUUGUCCAUCGCCUACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUA ..((((((...(((.((....)).)))....(((((((...........)))))))))))))((((......(....)...))))...(((.(((((((.......))))))).)))... ( -42.40) >Blich.0 96991 120 + 4222334/1360-1480 UGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUA ..((((((.........(((....)))....(((((((...........)))))))))))))((((......(....)...))))...(((.(((((((.......))))))).)))... ( -42.40) >Bsubt.0 92183 120 + 4214630/1360-1480 UGUUGUCCAUCGCCUACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUA ..((((((.........(((....)))....(((((((...........)))))))))))))((((......(....)...))))...(((.(((((((.......))))))).)))... ( -42.40) >consensus UGUUGUCCAUCGCCUACGCCUGUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCCUCUUUCGCUA ..((((((.........(((....)))....(((((((...........)))))))))))))((((......(....)...))))...(((.(((((((.......))))))).)))... (-42.50 = -42.00 + -0.50)

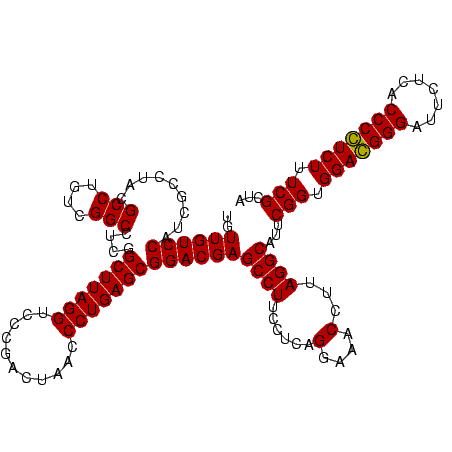
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.24 |
| Mean single sequence MFE | -37.40 |
| Consensus MFE | -35.30 |
| Energy contribution | -32.55 |
| Covariance contribution | -2.75 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.94 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.653331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1520-1640 GGAGGGACGCAGAASGAUAGAAGAAGCGUGCGAUUGGUUGUGCACGUCCAAGCAGUUAGGCUGAUAAGUAGGCAAAUCCGCUUAUCGUGAAGGCUGAGCUGUGAUGGGGAAGCUCCUUAU ..(((((.((...............(((((((........)))))))(((.((((((.(((((((((((.((.....))))))))).....)))).))))))..)))....))))))).. ( -38.20) >Banth.0 84076 120 + 5227293/1520-1640 GGAGGGACGCAGAAGGAUAGAAGAAGCGUGCGAUUGGUUGUGCACGUCCAAGCAGUUAGGCUGAUAAGUAGGCAAAUCCGCUUAUCGUGAAGGCUGAGCUGUGAUGGGGAAGCUCCUUAU ..(((((.((...............(((((((........)))))))(((.((((((.(((((((((((.((.....))))))))).....)))).))))))..)))....))))))).. ( -38.20) >Blich.0 96991 118 + 4222334/1520-1640 GGGGGGACGCAGGAGGAUAGGGUAAGCGCGGUAUUGGAUAUCCGCGUCCAAGCAGUUAGGCUGGGAAAUAGGCAAAUCCGUUUCCCGU-AAGGCUGAGCUGUGAUGGCGA-GCGAAAUAU .......(((...............((((((..........))))))(((.((((((.(((((((((((.((.....)))))))))..-..)))).))))))..)))...-)))...... ( -36.60) >Bsubt.0 92183 118 + 4214630/1520-1640 GGGGGGACGCAGGAGGAUAGGGUAAGCGCGGUAUUGGAUAUCCGCGUCCAAGCAGUUAGGCUGGGAAAUAGGCAAAUCCGUUUCCCAU-AAGGCUGAGCUGUGAUGGCGA-GCGAAAUAU .......(((...............((((((..........))))))(((.((((((.(((((((((((.((.....)))))))))..-..)))).))))))..)))...-)))...... ( -36.60) >consensus GGAGGGACGCAGAAGGAUAGAAGAAGCGCGCGAUUGGAUAUCCACGUCCAAGCAGUUAGGCUGAGAAAUAGGCAAAUCCGCUUACCGU_AAGGCUGAGCUGUGAUGGCGA_GCGAAAUAU ........((...............((((((.((.....))))))))(((.((((((.(((((((((((.((.....))))))))).....)))).))))))..)))....))....... (-35.30 = -32.55 + -2.75)

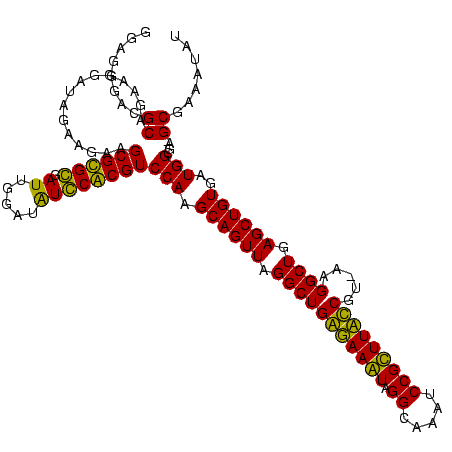
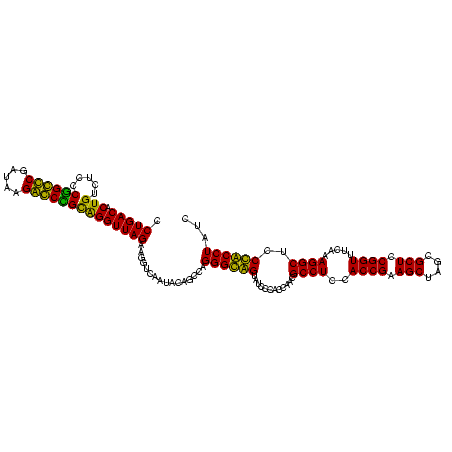
| Location | 84,077 – 84,186 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.86 |
| Mean single sequence MFE | -37.73 |
| Consensus MFE | -26.65 |
| Energy contribution | -29.52 |
| Covariance contribution | 2.88 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 109 + 5224283/1720-1840 GUAGGCCAGGAGAGAAUCCUAAGGUGUGCGAGAGAACUCUGGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUUUCUUAACG-----------GAAAGCCGCA (((.(((((((.....))))..))).)))..(((...(((......))).))).((......(((((....(....).....)))))(((((((....)-----------)))))).)). ( -35.10) >Banth.0 84076 109 + 5227293/1720-1840 GUAGGCGAGGAGAGAAUCCUAAGGUGUGCGAGAGAACUCUGGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUUUCUUAACG-----------GAAAGCCGCA (((.((.((((.....))))...)).)))..(((...(((......))).))).((......(((((....(....).....)))))(((((((....)-----------)))))).)). ( -32.80) >Blich.0 96991 120 + 4222334/1720-1840 GUAGGCGAGGAGAGAAUCCUAAGGUGAUCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCA ....(((....(((..((((....(((.(((((....)))))))))))).))).........(((((....(....).....)))))(((((.((.((((....)))))).)))))))). ( -41.50) >Bsubt.0 92183 120 + 4214630/1720-1840 GUAGGCGAGGAGAGAAUCCUAAGGUGAUCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCA ....(((....(((..((((....(((.(((((....)))))))))))).))).........(((((....(....).....)))))(((((.((.((((....)))))).)))))))). ( -41.50) >consensus GUAGGCGAGGAGAGAAUCCUAAGGUGAGCGAGAGAACUCUCGUUAAGGAACUCGGCAAAAUGACCCCGUAACUUCGGGAGAAGGGGUGCUCUCUUAACG___________GAAAGCCGCA ....(((....(((..((((..((.((((....)))).)).....)))).))).........(((((.....(((....))))))))((((((...((((....))))..))))))))). (-26.65 = -29.52 + 2.88)

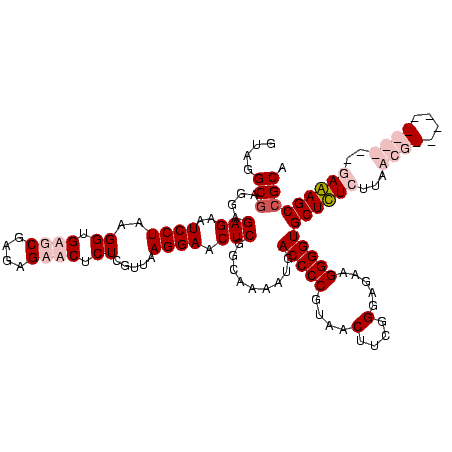
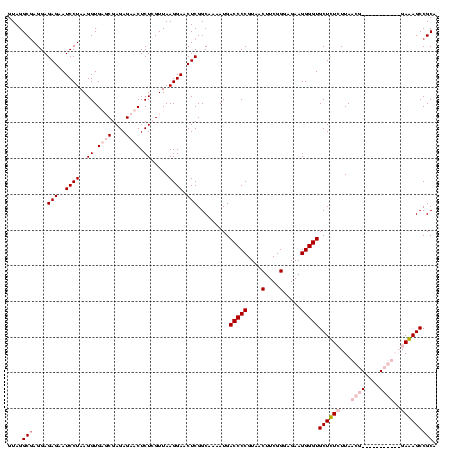
| Location | 84,077 – 84,186 |
|---|---|
| Length | 109 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.72 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -31.83 |
| Energy contribution | -34.95 |
| Covariance contribution | 3.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.665130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 109 + 5224283/1800-1920 AAGGGGUGCUUUCUUAACG-----------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUG ...(((((((((((....)-----------)))))).((..(((((((((....))..))))))))).....(((..((((.(((...(((....)))...))).)))).)))..)))). ( -34.70) >Banth.0 84076 109 + 5227293/1800-1920 AAGGGGUGCUUUCUUAACG-----------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUG ...(((((((((((....)-----------)))))).((..(((((((((....))..))))))))).....(((..((((.(((...(((....)))...))).)))).)))..)))). ( -34.70) >Blich.0 96991 120 + 4222334/1800-1920 AAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUG ...(((((((((.((.((((....)))))).)))))............))))(((((..(((((.....)))))..(((((((((...(((....)))...))..)))))))..))))). ( -45.10) >Bsubt.0 92183 120 + 4214630/1800-1920 AAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUG ...(((((((((.((.((((....)))))).)))))............))))(((((..(((((.....)))))..(((((((((...(((....)))...))..)))))))..))))). ( -45.10) >consensus AAGGGGUGCUCUCUUAACG___________GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACACCUG .(((.((((((((...((((....))))..)))))).((..(((((((((....))..))))))))).........(((((((((...(((....)))...))..))))))).)).))). (-31.83 = -34.95 + 3.12)

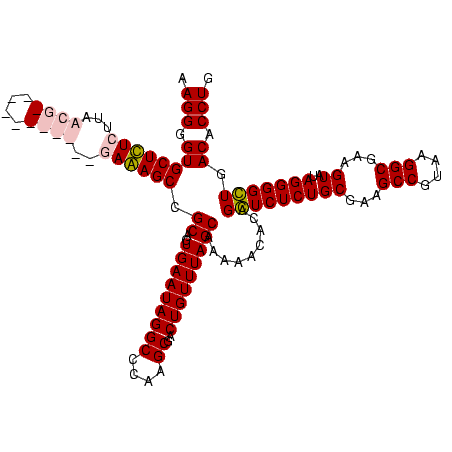
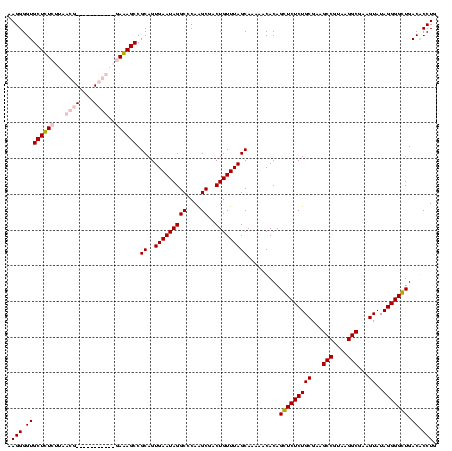
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -46.75 |
| Consensus MFE | -41.50 |
| Energy contribution | -44.25 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.936982 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1880-2000 UCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACU ..(((...(((....)))...)))((.(((....)..(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))))).))... ( -45.50) >Banth.0 84076 120 + 5227293/1880-2000 UCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACU ..(((...(((....)))...)))((.(((....)..(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))))).))... ( -45.50) >Blich.0 96991 120 + 4222334/1880-2000 UCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACU ..((((..(((...(((((.(((......)))..)))))(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))))))))... ( -48.00) >Bsubt.0 92183 120 + 4214630/1880-2000 UCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACU ..((((..(((...(((((.(((......)))..)))))(((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))))))))... ( -48.00) >consensus UCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAAGAGCGCUUAGCGUAAGCGAAGCUCCGAACUGAAGCCCCAGUAAACGGCGGCCGUAACU ..(((...(((....)))...)))....((...(((.(((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...))))).)))..)) (-41.50 = -44.25 + 2.75)

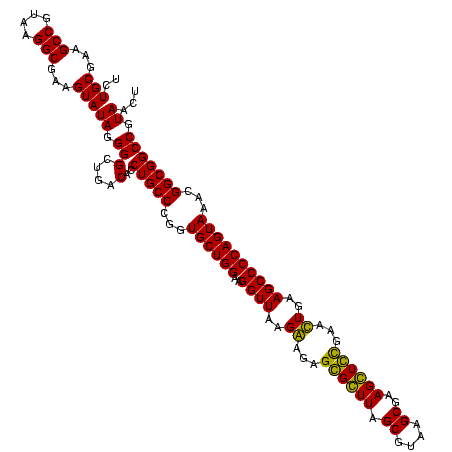
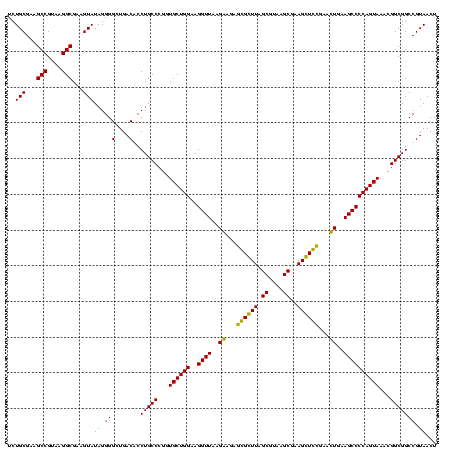
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.56 |
| Mean single sequence MFE | -40.65 |
| Consensus MFE | -40.40 |
| Energy contribution | -40.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/1920-2040 CCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGU (((..((((((..((((..((...((((((.((....))..))))))...))..))))))))))...(((((((((.......))))))..((((..........))))))))))..... ( -39.90) >Banth.0 84076 120 + 5227293/1920-2040 CCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGU (((..((((((..((((..((...((((((.((....))..))))))...))..))))))))))...(((((((((.......))))))..((((..........))))))))))..... ( -39.90) >Blich.0 96991 120 + 4222334/1920-2040 CCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGU (((..((((((..((((..((...((((((.((....))..))))))...))..))))))))))...(((((((((.......))))))..((((..........))))))))))..... ( -41.40) >Bsubt.0 92183 120 + 4214630/1920-2040 CCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGU (((..((((((..((((..((...((((((.((....))..))))))...))..))))))))))...(((((((((.......))))))..((((..........))))))))))..... ( -41.40) >consensus CCCGGUGCUGGAAGGUUAAGAAGAGCGCUUAGCGUAAGCGAAGCUCCGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUCCUUGUCGGGUAAGU (((..((((((..((((..((...((((((.((....))..))))))...))..))))))))))...(((((((((.......))))))..((((..........))))))))))..... (-40.40 = -40.65 + 0.25)

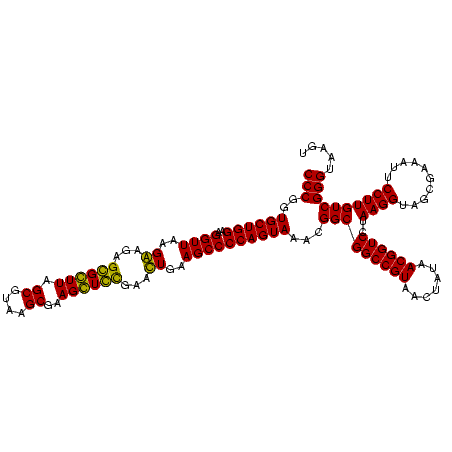
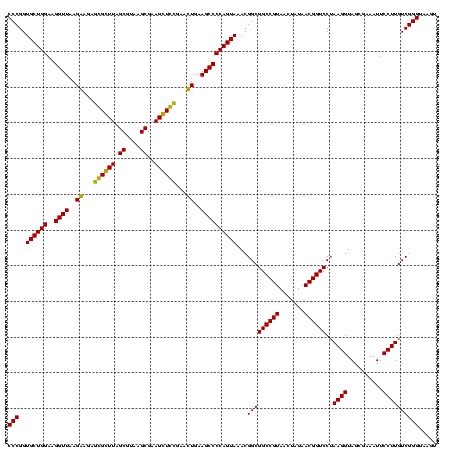
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -45.95 |
| Consensus MFE | -45.75 |
| Energy contribution | -44.25 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.67 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.997977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2160-2280 UACUGUAGCCUGAUAUUGAAUUUUGGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUU ..................(((((..(((((((.....((((.....(((..(((((....((((((((....)))))))).))))).)))((((....)))))))))))))))..))))) ( -46.00) >Banth.0 84076 120 + 5227293/2160-2280 UACUGUAGCCUGAUAUUGAAUUUUGGUACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUU ..................(((((..(((((((.....((((.....(((..(((((....((((((((....)))))))).))))).)))((((....)))))))))))))))..))))) ( -46.00) >Blich.0 96991 120 + 4222334/2160-2280 UACUGCAGCCUGAUAUUGAAUGUUGGUACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUU ...(((((((.(((.(((..((((.((((.....))))..))))..)))..(((((....((((((((....)))))))).))))))))(((((....)))))..)))))))........ ( -45.90) >Bsubt.0 92183 120 + 4214630/2160-2280 UACUGCAGCCUGAUAUUGAAUGUUGGUACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUU ...(((((((.(((.(((..((((.((((.....))))..))))..)))..(((((....((((((((....)))))))).))))))))(((((....)))))..)))))))........ ( -45.90) >consensus UACUGCAGCCUGAUAUUGAAUGUUGGUACAGCUUGUACAGGAUAGGCAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCACCGGUGGGAUACCACCCUGACUGUAUUGAAAUU ..................(((((..(((((((.....(((........((.(((((....((((((((....)))))))).))))).))(((((....)))))))))))))))..))))) (-45.75 = -44.25 + -1.50)

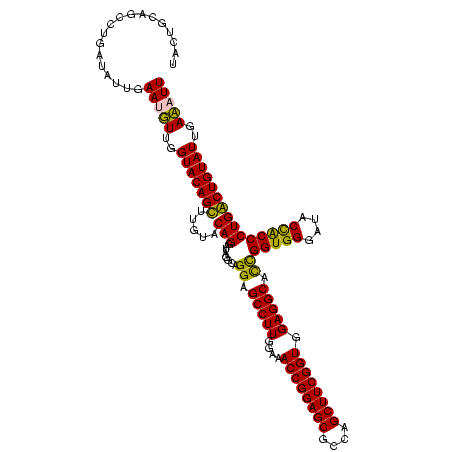
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.67 |
| Mean single sequence MFE | -33.47 |
| Consensus MFE | -32.73 |
| Energy contribution | -31.23 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991798 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2160-2280 AAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGUACCAAAAUUCAAUAUCAGGCUACAGUA ........(((((...(((((((.....)))...((((..((((.(((....))).)))).....))))....))))...)))))...((((((((..............)).)))))). ( -33.74) >Banth.0 84076 120 + 5227293/2160-2280 AAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGUACCAAAAUUCAAUAUCAGGCUACAGUA ........(((((...(((((((.....)))...((((..((((.(((....))).)))).....))))....))))...)))))...((((((((..............)).)))))). ( -33.74) >Blich.0 96991 120 + 4222334/2160-2280 AAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCUGUACAAGCUGUACCAACAUUCAAUAUCAGGCUGCAGUA ..........((((((((((((.(((.....)))((((.....((((((((....))))))))..))))......)))))))((((.....))))...............)))))..... ( -33.20) >Bsubt.0 92183 120 + 4214630/2160-2280 AAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCUGUACAAGCUGUACCAACAUUCAAUAUCAGGCUGCAGUA ..........((((((((((((.(((.....)))((((.....((((((((....))))))))..))))......)))))))((((.....))))...............)))))..... ( -33.20) >consensus AAGGUCAAUACAGCCAGGGCAGUAUCCCACCAACGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCACCUAUCCUGUACAAACUGUACCAAAAUUCAAUAUCAGGCUACAGUA ........(((((...((((((............((((..((((.(((....))).)))).....))))..))))))...)))))...((((((((..............)).)))))). (-32.73 = -31.23 + -1.50)


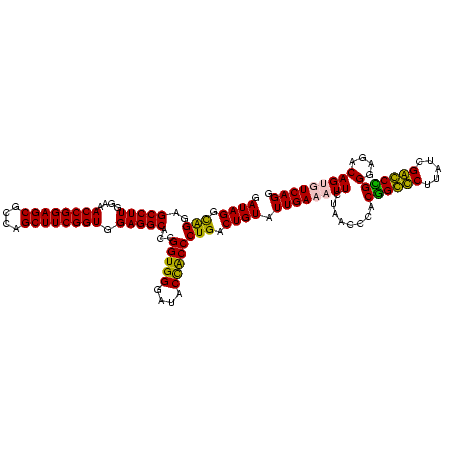
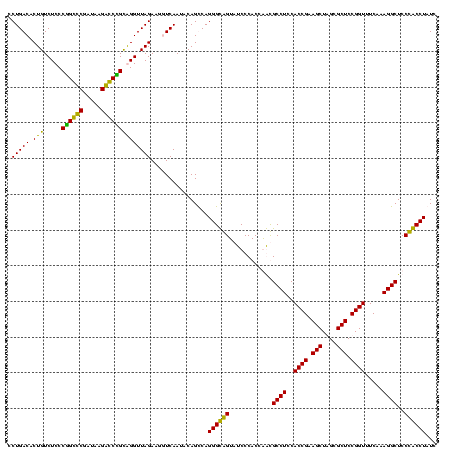
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -52.67 |
| Consensus MFE | -52.45 |
| Energy contribution | -50.82 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.51 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.997381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2200-2320 GAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGG ......(((..(((((....((((((((....)))))))).))))).)))((((....))))((((((....................((((((.....))))))(....)...)))))) ( -50.00) >Banth.0 84076 120 + 5227293/2200-2320 GAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGG ......(((..(((((....((((((((....)))))))).))))).)))((((....))))((((((....................((((((.....))))))(....)...)))))) ( -50.00) >Blich.0 96991 120 + 4222334/2200-2320 GAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGG .((((.((((((((((....((((((((....)))))))).))))).))(((((....)))))))).)))).((((((((((...((.((((((.....))))))))))).)).))))). ( -54.60) >Bsubt.0 92183 120 + 4214630/2200-2320 GAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGG .((((.((((((((((....((((((((....)))))))).))))).))(((((....)))))))).)))).((((((((((...((.((((((.....))))))))))).)).))))). ( -56.10) >consensus GAUAGGCAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCACCGGUGGGAUACCACCCUGACUGUAUUGAAAUUCUAACCCACGGCCCUUAUCGACCCGGGAGACAGUGUCAGG .((((.(((..(((((....((((((((....)))))))).)))))...(((((....)))))))).)))).((((((((........((((((.....))))))(....))))))))). (-52.45 = -50.82 + -1.63)

| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.61 |
| Mean single sequence MFE | -42.57 |
| Consensus MFE | -40.09 |
| Energy contribution | -36.97 |
| Covariance contribution | -3.13 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971966 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2200-2320 CCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUC .(((((.(((.((..((((((.....))))))..)).))).((....))...)))))((((((.....)))...((((..((((.(((....))).)))).....))))....))).... ( -40.30) >Banth.0 84076 120 + 5227293/2200-2320 CCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUC .(((((.(((.((..((((((.....))))))..)).))).((....))...)))))((((((.....)))...((((..((((.(((....))).)))).....))))....))).... ( -40.30) >Blich.0 96991 120 + 4222334/2200-2320 CCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUC ..((((.(((.(((.((((((.....)))))).))).)))...))))....((((..((..(((((.....)))))..))...((((((((....))))))))...)))).......... ( -44.10) >Bsubt.0 92183 120 + 4214630/2200-2320 CCUGACACUGUCUCCCCGCCCGAUAAGGGCGGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUC ..((((.(((.(((.((((((.....)))))).))).)))...))))....((((..((..(((((.....)))))..))...((((((((....))))))))...)))).......... ( -45.60) >consensus CCUGACACUGUCUCCCGGCCCGAUAAGACCCGCAGGUUAGAAGGUCAAUACAGCCAGGGCAGUAUCCCACCAACGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCACCUAUC .(((((.(((.....((((((.....))))))))))))))................((((((............((((..((((.(((....))).)))).....))))..))))))... (-40.09 = -36.97 + -3.13)

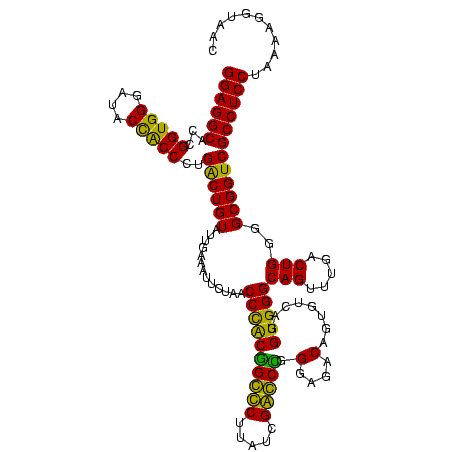
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -49.98 |
| Consensus MFE | -51.00 |
| Energy contribution | -48.12 |
| Covariance contribution | -2.88 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.90 |
| Structure conservation index | 1.02 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2240-2360 GGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAAC (((((((..........((((((((((((((.((((....((......((((((.....))))))(....)))..))))....)))))).....)))))))))))))))........... ( -49.30) >Banth.0 84076 120 + 5227293/2240-2360 GGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAAC (((((((..........((((((((((((((.((((....((......((((((.....))))))(....)))..))))....)))))).....)))))))))))))))........... ( -49.30) >Blich.0 96991 120 + 4222334/2240-2360 GGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAAC ((((((...(((((....)))))..((((((.((((((((((...((.((((((.....))))))))))).)).)))))....)))))).(((((...)))))))))))........... ( -49.90) >Bsubt.0 92183 120 + 4214630/2240-2360 GGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAAC ((((((...(((((....)))))..((((((.((((((((((...((.((((((.....))))))))))).)).)))))....)))))).(((((...)))))))))))........... ( -51.40) >consensus GGAGGCACCGGUGGGAUACCACCCUGACUGUAUUGAAAUUCUAACCCACGGCCCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAAC ((((((...(((((....)))))..((((((.............((((((((((.....))))).(....)........)))))(((.....)))..))))))))))))........... (-51.00 = -48.12 + -2.88)

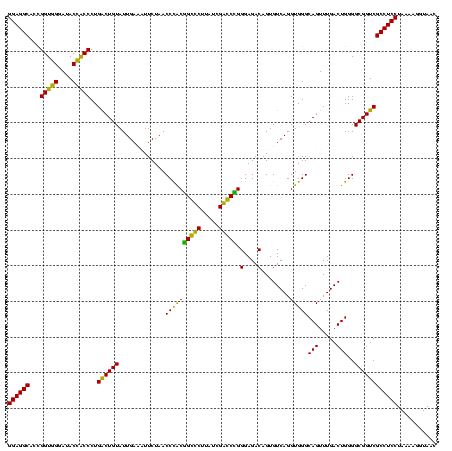
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.72 |
| Mean single sequence MFE | -45.12 |
| Consensus MFE | -43.45 |
| Energy contribution | -41.83 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.96 |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.998018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2240-2360 GUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCC ...........((((((((((((((..((((...........)))).(((.((..((((((.....))))))..)).)))................)))))))..........))))))) ( -44.00) >Banth.0 84076 120 + 5227293/2240-2360 GUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCC ...........((((((((((((((..((((...........)))).(((.((..((((((.....))))))..)).)))................)))))))..........))))))) ( -44.00) >Blich.0 96991 120 + 4222334/2240-2360 GUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCC ...........(((((((((.......)))..(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))...........)))))) ( -45.50) >Bsubt.0 92183 120 + 4214630/2240-2360 GUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCCGCCCGAUAAGGGCGGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCC ...........(((((((((.......)))..(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))...........)))))) ( -47.00) >consensus GUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGCCCGAUAAGACCCGCAGGUUAGAAGGUCAAUACAGCCAGGGCAGUAUCCCACCAACGCCUCC ...........(((((((((.......)))..(((((((..(((((.(((.....((((((.....))))))))))))))..(((.......))).)))))))...........)))))) (-43.45 = -41.83 + -1.63)

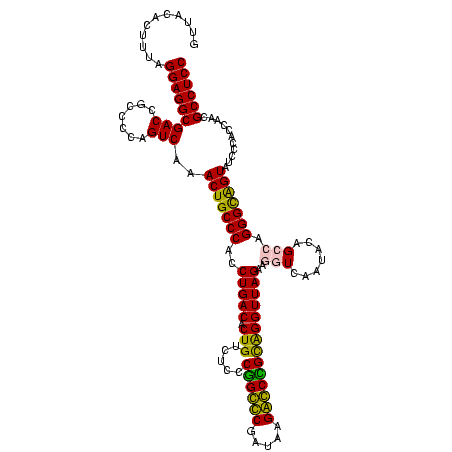
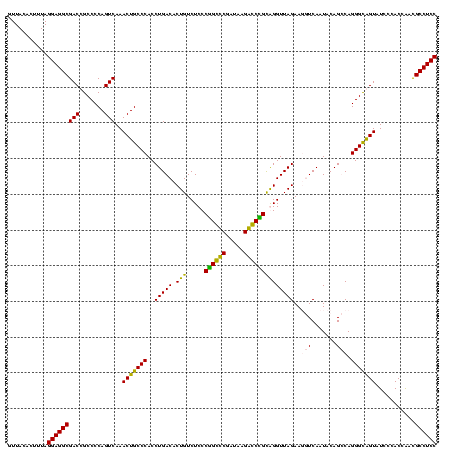
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -48.82 |
| Consensus MFE | -50.15 |
| Energy contribution | -48.52 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.11 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986071 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2280-2400 CUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAU ((((((...(((((.....)))))(((((((...)))...(((((.....(((((...)))))((((((...........))))))))))).....)))).......))))))....... ( -48.80) >Banth.0 84076 120 + 5227293/2280-2400 CUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAU ((((((...(((((.....)))))(((((((...)))...(((((.....(((((...)))))((((((...........))))))))))).....)))).......))))))....... ( -48.80) >Blich.0 96991 120 + 4222334/2280-2400 CUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAU ((((((.(((((((..(((.((((.(....)..)))).))))))))))....((((((.((.(((((((...........))))))).))........))))))...))))))....... ( -48.80) >Bsubt.0 92183 120 + 4214630/2280-2400 CUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAU ((((((...(((((.....)))))(((((((...)))...(((((.....(((((...)))))((((((...........))))))))))).....)))).......))))))....... ( -48.90) >consensus CUAACCCACGGCCCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAU ((((((...(((((.....)))))(((((((...)))...(((((.....(((((...)))))((((((...........))))))))))).....)))).......))))))....... (-50.15 = -48.52 + -1.63)

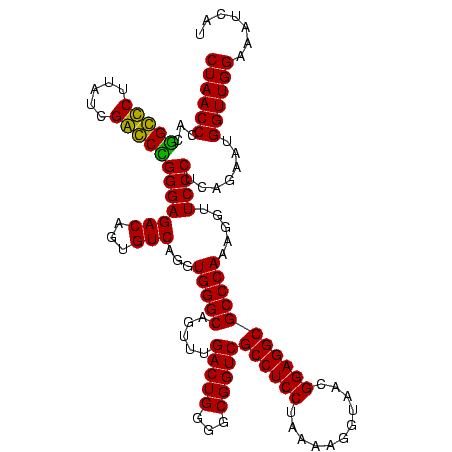
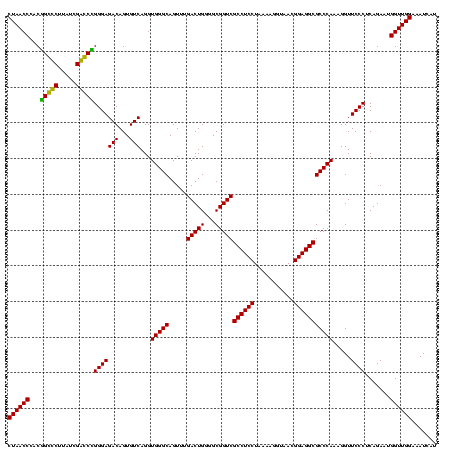
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.17 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -45.20 |
| Energy contribution | -43.57 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.21 |
| Structure conservation index | 1.03 |
| SVM decision value | 2.22 |
| SVM RNA-class probability | 0.990516 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2280-2400 AUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAG .........((((.......((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....)))))...)))).. ( -44.00) >Banth.0 84076 120 + 5227293/2280-2400 AUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAG .........((((.......((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....)))))...)))).. ( -44.00) >Blich.0 96991 120 + 4222334/2280-2400 AUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAG .........((((......(((((.....(((((((((((...........))))))(((.......))).....)))))...(((...))))))))((((.....))))....)))).. ( -43.10) >Bsubt.0 92183 120 + 4214630/2280-2400 AUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCCGCCCGAUAAGGGCGGCGGGUUAG .........((((.......((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....)))))...)))).. ( -43.70) >consensus AUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGCCCGAUAAGACCCGCAGGUUAG .........((((.......((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....)))))...)))).. (-45.20 = -43.57 + -1.63)

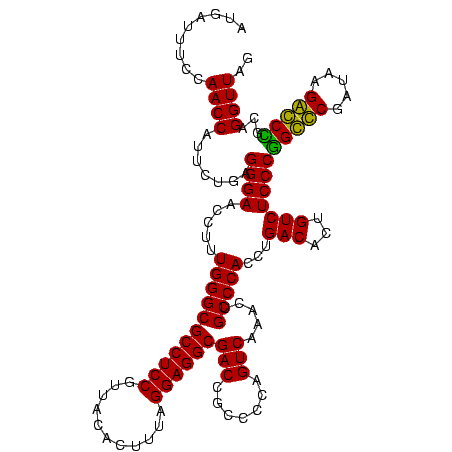

| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -47.55 |
| Consensus MFE | -48.05 |
| Energy contribution | -47.55 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.87 |
| Structure conservation index | 1.01 |
| SVM decision value | 2.11 |
| SVM RNA-class probability | 0.988310 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2320-2440 UGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGAC ...((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..... ( -47.30) >Banth.0 84076 120 + 5227293/2320-2440 UGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGAC ...((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..... ( -47.30) >Blich.0 96991 120 + 4222334/2320-2440 UGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGAC ...((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..... ( -47.80) >Bsubt.0 92183 120 + 4214630/2320-2440 UGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGAC ...((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..... ( -47.80) >consensus UGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGAC ...((((((......((((....((((((...........))))))))))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))))))))..... (-48.05 = -47.55 + -0.50)

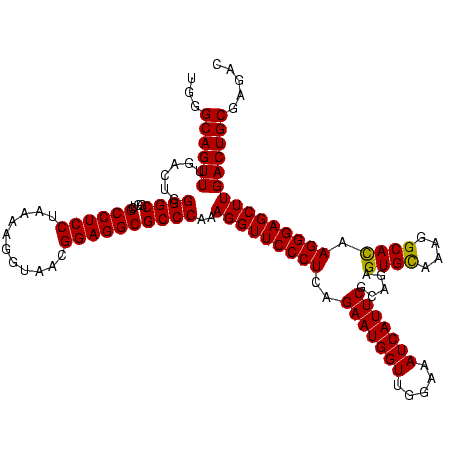
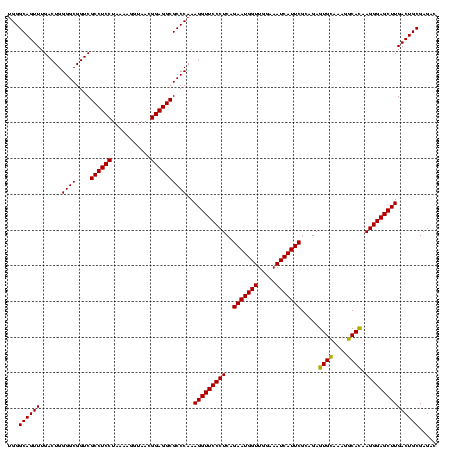
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -35.60 |
| Energy contribution | -36.10 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944399 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2320-2440 GUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCA .....(((((...((((((((((((....)))......(((((.........)))))))))))).....(((((((((((...........))))))).....))))))...)))))... ( -38.00) >Banth.0 84076 120 + 5227293/2320-2440 GUCUCGCAGUCAAGCUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCA .....(((((...((((((((((((....)))......(((((.........)))))))))))).....(((((((((((...........))))))).....))))))...)))))... ( -38.00) >Blich.0 96991 120 + 4222334/2320-2440 GUCUCGCAGUCAAGCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCA .....(((((...(((((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))).....))))))...)))))... ( -37.40) >Bsubt.0 92183 120 + 4214630/2320-2440 GUCUCGCAGUCAAGCUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCA .....(((((...(((((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))).....))))))...)))))... ( -37.40) >consensus GUCUCGCAGUCAAGCUCCCUUAUGCCUUUACACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCA .....(((((...(((((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))).....))))))...)))))... (-35.60 = -36.10 + 0.50)

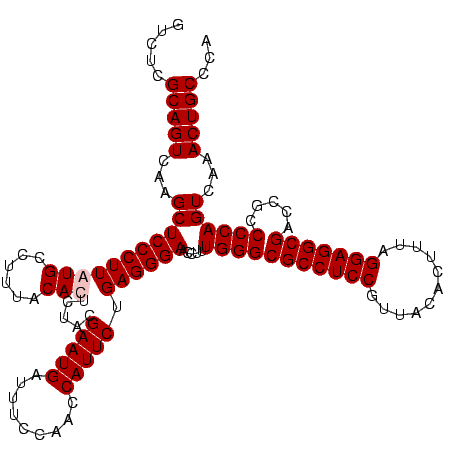
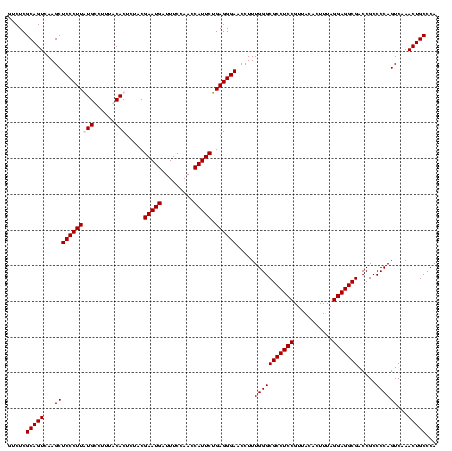
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -48.05 |
| Consensus MFE | -48.55 |
| Energy contribution | -48.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.68 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974282 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2360-2480 GGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCC (((.((((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))...))..))) ( -47.80) >Banth.0 84076 120 + 5227293/2360-2480 GGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCC (((.((((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))...))..))) ( -47.80) >Blich.0 96991 120 + 4222334/2360-2480 GGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCC (((.((((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))...))..))) ( -48.30) >Bsubt.0 92183 120 + 4214630/2360-2480 GGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCC (((.((((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))...))..))) ( -48.30) >consensus GGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCC (((.((((((..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....)))))))...))..))) (-48.55 = -48.05 + -0.50)

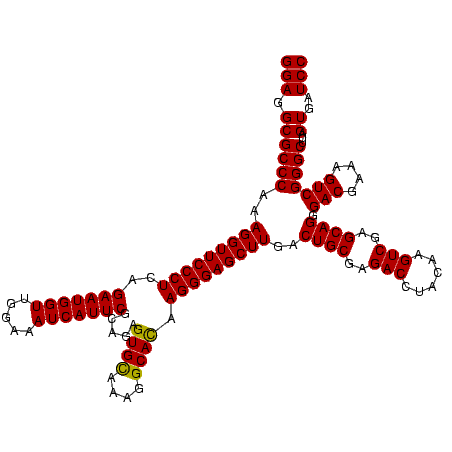
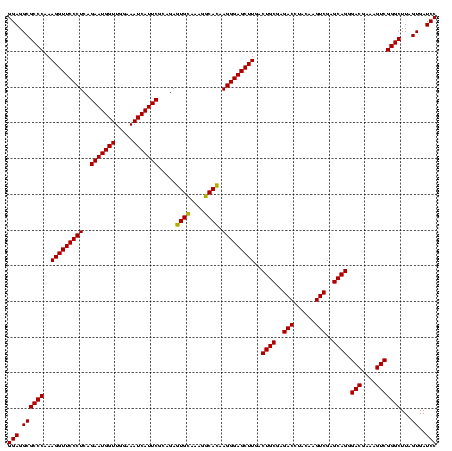
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -43.85 |
| Consensus MFE | -43.35 |
| Energy contribution | -43.60 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.32 |
| SVM RNA-class probability | 0.941912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2400-2520 UCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAA ((((.(((((....(((......((((((..((((..(((......)))..)))).(((....))))))))).....(((((((....))).))))...)))..))))).))))...... ( -43.80) >Banth.0 84076 120 + 5227293/2400-2520 UCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAA ((((.(((((....(((......((((((..((((..(((......)))..)))).(((....))))))))).....(((((((....))).))))...)))..))))).))))...... ( -43.80) >Blich.0 96991 120 + 4222334/2400-2520 UCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAA (((..(((((....(((.((...((((((..((((..(((......)))..)))).(((....)))))))))..)).(((((((....))).))))...)))..)))))..)))...... ( -43.90) >Bsubt.0 92183 120 + 4214630/2400-2520 UCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAA (((..(((((....(((.((...((((((..((((..(((......)))..)))).(((....)))))))))..)).(((((((....))).))))...)))..)))))..)))...... ( -43.90) >consensus UCGCAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCGCUCAACGGAUAAAA ((((.(((((....(((.((...((((((..((((..(((......)))..)))).(((....)))))))))..)).(((((((....))).))))...)))..))))).))))...... (-43.35 = -43.60 + 0.25)

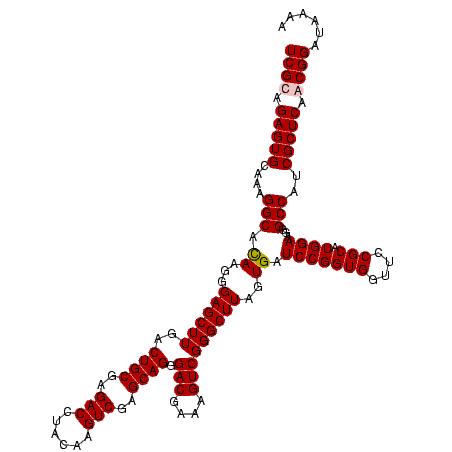
| Location | 84,077 – 84,197 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -54.80 |
| Consensus MFE | -54.80 |
| Energy contribution | -54.80 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.22 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.779112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 120 + 5224283/2560-2680 ACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCG ....(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..))....))))) ( -54.80) >Banth.0 84076 120 + 5227293/2560-2680 ACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCG ....(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..))....))))) ( -54.80) >Blich.0 96991 120 + 4222334/2560-2680 ACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCG ....(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..))....))))) ( -54.80) >Bsubt.0 92183 120 + 4214630/2560-2680 ACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCG ....(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..))....))))) ( -54.80) >consensus ACAUCGACGGGGAGGUUUGGCACCUCGAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCG ....(((((..(((((.....)))))((..((((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....)))))))..))....))))) (-54.80 = -54.80 + 0.00)

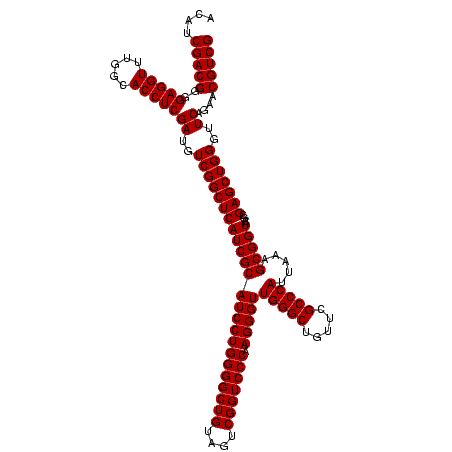
| Location | 84,077 – 84,196 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.54 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -30.25 |
| Energy contribution | -29.25 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.98 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 119 + 5224283/2800-2920 CUAUCAACCUGAUCAUCUUUCAGGAAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCUUCCGCACAUAGCUACCCA .......(((((.......)))))........(((((...-((.(((((....)))))...(((((((((.....)))(((.(((.....))).))).)))))).))....))))).... ( -29.70) >Banth.0 84076 119 + 5227293/2800-2920 CUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCUUCCGCACAUAGCUACCCA .......(((((.......)))))........(((((...-((.(((((....)))))...(((((((((.....)))(((.(((.....))).))).)))))).))....))))).... ( -29.90) >Blich.0 96991 120 + 4222334/2800-2920 CUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUAUAGGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCGUCCGCACAUAGCUACCCA ...(((((((((.......)))))..(((((.((((.....))))))))).........))))(((..(((...(((.(((.(((.....))).)))........)))...)))..))). ( -29.80) >Bsubt.0 92183 119 + 4214630/2800-2920 CUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUGC-GGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCGUCCGCACAUAGCUACCCA ((((...(((((.......))))).............(((-(((.(((((...(((.....)))(..(((.((.......)).)))..)........))))))))))).))))....... ( -34.60) >consensus CUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUACCUUAC_GCAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCACUUAUCCCGUCCGCACAUAGCUACCCA ...(((((((((.......)))))..(((((.((((.....))))))))).........))))(((..(((...(((.(((.(((.....))).)))........)))...)))..))). (-30.25 = -29.25 + -1.00)

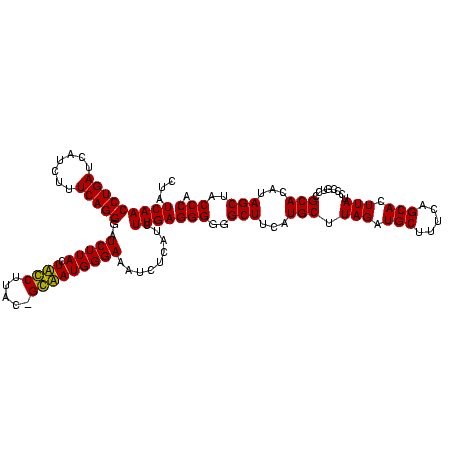
| Location | 84,077 – 84,196 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -31.06 |
| Energy contribution | -30.62 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.640743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 119 + 5224283/2840-2960 UAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUAGC-GUAAGCUAGUAAGAUUCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAU ..(((........((((((.......(((((..(.((((-....)))))..)))))(((((.......)))))...........)))).))..(((((....)))))...)))....... ( -33.30) >Banth.0 84076 119 + 5227293/2840-2960 UAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUAGC-GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAU ..(((........((((((.......(((((..(.((((-....)))))..)))))(((((.......)))))...........)))).))..(((((....)))))...)))....... ( -35.70) >Blich.0 96991 120 + 4222334/2840-2960 UAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUUCCUAUAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAU ..(((........(((((((.(((..(((((..(.((((.....)))))..)))))(((((.......))))).......)))))))).))..(((((....)))))...)))....... ( -34.30) >Bsubt.0 92183 119 + 4214630/2840-2960 UAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUUCC-GCAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAU ..(((........(((((((.(((..(((((..(.((((-....)))))..)))))(((((.......))))).......)))))))).))..(((((....)))))...)))....... ( -37.80) >consensus UAAGCAUGAAGCCCCCCUCAAGAUGAGAUUUCCCAUACC_GUAAGCAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCCGAGGUGGAAGCAUGGCGACACAUGGAGCUGACAAAU ..(((........(((((((.(((..(((((..(.((((.....)))))..)))))(((((.......))))).......)))))))).))..(((((....)))))...)))....... (-31.06 = -30.62 + -0.44)

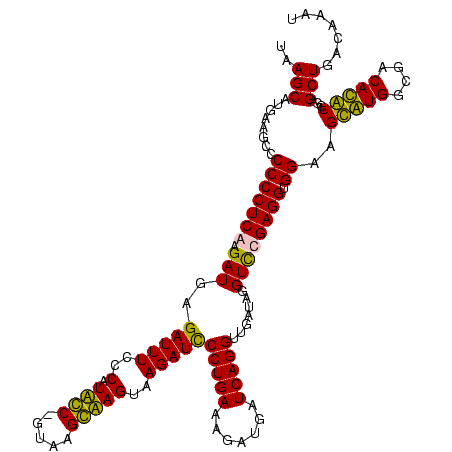
| Location | 84,077 – 84,196 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -32.73 |
| Consensus MFE | -34.80 |
| Energy contribution | -32.30 |
| Covariance contribution | -2.50 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.38 |
| Structure conservation index | 1.06 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997891 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 119 + 5224283/2840-2960 AUUCGUCAGCUCCACAUGUCACCAUGCUUCCACCUCGAACCUAUCAACCUGAUCAUCUUUCAGGAAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUA .......((((((.((((....))))......((((((.........(((((.......)))))..(((((.((((....-))))))))).........))))))))))))......... ( -32.30) >Banth.0 84076 119 + 5227293/2840-2960 AUUCGUCAGCUCCACAUGUCACCAUGCUUCCACCUCGAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUA .......((((((.((((....))))......((((((.........(((((.......)))))..(((((.((((....-))))))))).........))))))))))))......... ( -32.50) >Blich.0 96991 120 + 4222334/2840-2960 AUCUGUCAGCUCCAUGUGUCGCCACACUUCCACCUCAGACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUAUAGGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUA .......((((((..((((....)))).....((((((.........(((((.......)))))..(((((.((((.....))))))))).........))))))))))))......... ( -33.20) >Bsubt.0 92183 119 + 4214630/2840-2960 AUCUGUCAGCUCCAUGUGUCGCCACACUUCCACCUCAGACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUGC-GGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUA .......((((((..((((....)))).....((((((.........(((((.......)))))..(((((.((((....-))))))))).........))))))))))))......... ( -32.90) >consensus AUCCGUCAGCUCCACAUGUCACCACACUUCCACCUCAAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUACCUUAC_GCAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUA .......((((((.((((....))))......((((((.........(((((.......)))))..(((((.((((.....))))))))).........))))))))))))......... (-34.80 = -32.30 + -2.50)

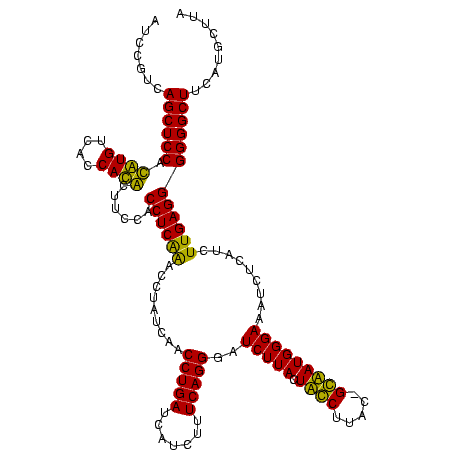
| Location | 84,077 – 84,188 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.52 |
| Mean single sequence MFE | -28.93 |
| Consensus MFE | -27.44 |
| Energy contribution | -27.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878415 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 84077 111 + 5224283/2880-3000 GUAAGCUAGUAAGAUUCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC---AUAU-----AAU-A .((((((.....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))..)).))))...---....-----...-. ( -25.60) >Banth.0 84076 111 + 5227293/2880-3000 GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC---AUAU-----AAU-A .((((((.....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))..)).))))...---....-----...-. ( -28.00) >Blich.0 96991 120 + 4222334/2880-3000 AUAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACCUAAAUAUUUGUGAAUCG ...(((((((..(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))....))))..)))................ ( -31.40) >Bsubt.0 92183 117 + 4214630/2880-3000 GCAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACC---AUAUUUUUGAAUGA ....((((((..(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))....))))..))---.............. ( -30.70) >consensus GUAAGCAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCCGAGGUGGAAGCAUGGCGACACAUGGAGCUGACAAAUACUAAUAGAUCGAGGACUUAACC___AUAU_____AAU_A ......((((..(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))....))))..................... (-27.44 = -27.25 + -0.19)

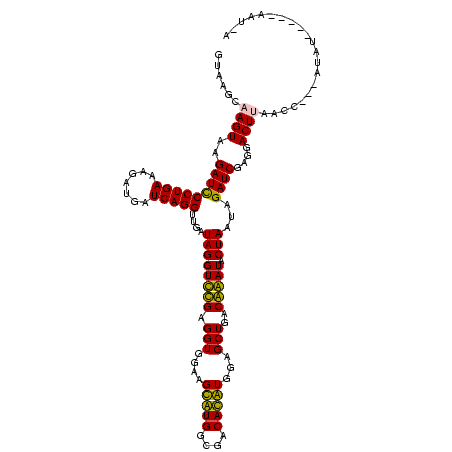
Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:23:16 2006