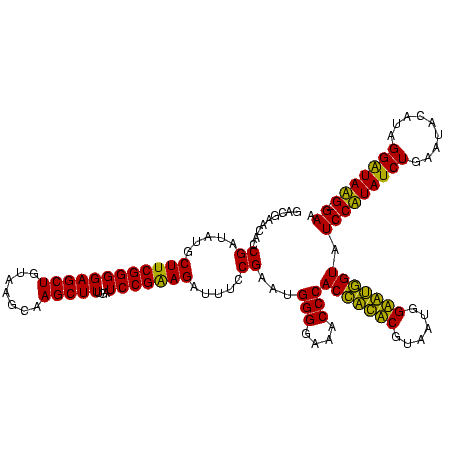
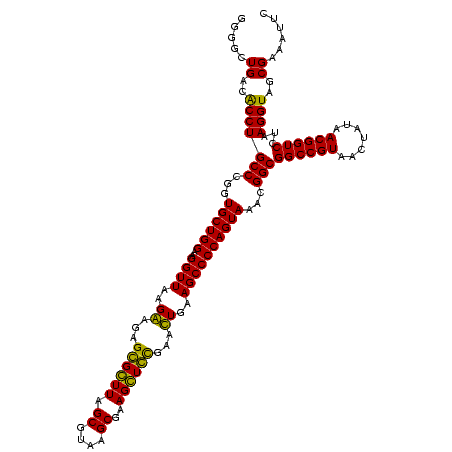
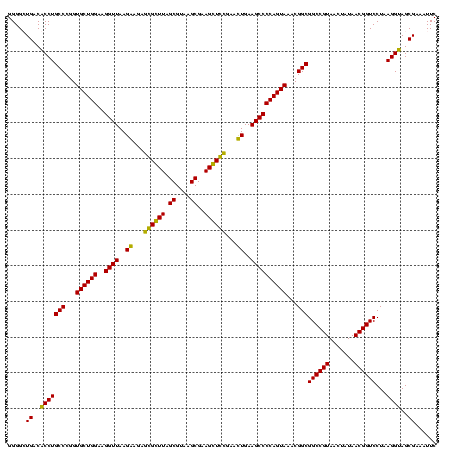
| Sequence ID | Blich.0 |
|---|---|
| Location | 36,508 – 36,628 |
| Length | 120 |
| Max. P | 0.999308 |

| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -41.75 |
| Consensus MFE | -42.44 |
| Energy contribution | -39.75 |
| Covariance contribution | -2.69 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.49 |
| Structure conservation index | 1.02 |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.999308 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/40-160 GACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAA ........((((......))))((((((.......))))))....(((......)))...(((....)))(((((((......))))))).(((((((((.........))))))).)). ( -41.00) >Bsubt.0 32193 120 + 4214630/40-160 GACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAA ........((((......))))((((((.......))))))....(((......)))...(((....)))(((((((......))))))).(((((((((.........))))))).)). ( -41.00) >Bcere.0 31024 119 + 5224283/40-160 GACUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAA .........((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........)))))))-)). ( -42.20) >Banth.0 31045 119 + 5227293/40-160 GACUAACGCCGAUAUGCUUCGGGGAGCUGUAAGUAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGAUAAG-GAA .........((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........)))))))-)). ( -42.80) >consensus GACGAACACCGAUAUGCUUCGGGGAGCUGUAAGCAAGCUUUGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCACACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAAG_GAA .........((.....((((((((((((.......)))))...))))))).....))...(((....)))(((((((......))))))).(((((((((.........))))))).)). (-42.44 = -39.75 + -2.69)

| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -33.08 |
| Consensus MFE | -29.77 |
| Energy contribution | -27.53 |
| Covariance contribution | -2.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.81 |
| SVM RNA-class probability | 0.978213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/40-160 UUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUUCGUC ............((((((...((((((...(((((((......)))))))(((.......(((((((....)))))))...((((.......)))).)))....))))))...)))))). ( -34.60) >Bsubt.0 32193 120 + 4214630/40-160 UUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUGCUUACAGCUCCCCGAAGCAUAUCGGUGUUCGUC ............((((((...((((((...(((((((......)))))))(((.......(((((((....)))))))...((((.......)))).)))....))))))...)))))). ( -34.60) >Bcere.0 31024 119 + 5224283/40-160 UUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUAGUC ...-((.(((((((((..........(((.(((.((((((.....))))))))).)))...(((((..(((....)))...((((.......))))..))))))))))..)).)).)).. ( -30.40) >Banth.0 31045 119 + 5227293/40-160 UUC-CUUAUCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUAGUC .((-((((((...........))))))))((((((((......))))))))(((((((......)))))))(((((.....((((.......))))..)))))((......))....... ( -32.70) >consensus UUC_CAUAUCCUAUGAAUUCAGAUAAGGAUACCACACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCAAAGCUUACUUACAGCUCCCCGAAGCAUAUCGGCGUUAGUC .((.((((((...((....)))))))))).(((((((......)))))))(((....)))(((((((....)))))))...((((.......))))..((((......))))........ (-29.77 = -27.53 + -2.25)


| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -40.90 |
| Consensus MFE | -40.09 |
| Energy contribution | -37.15 |
| Covariance contribution | -2.94 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.64 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.27 |
| SVM RNA-class probability | 0.998893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/80-200 UGAUCCGGAGAUUUCCGAAUGGGGAAACCCACUGCUCGUAAUGGAGCAGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGA .....(((......)))...(((....)))(((((((......))))))).(((...((((..(((.((.....)).)))..)))).(((((..(((..........))).))))).))) ( -42.90) >Bsubt.0 32193 120 + 4214630/80-200 UGAUCCGGAGAUUUCCGAAUGGGGAAACCCACCACUCGUAAUGGAGUGGUAUCCAUAUCUGAAUUCAUAGGAUAUGAGAAGGCAGACCCGGGGAACUGAAACAUCUAAGUACCCGGAGGA .....(((......)))...(((....)))(((((((......))))))).(((...((((..(((.((.....)).)))..)))).(((((..(((..........))).))))).))) ( -42.90) >Bcere.0 31024 119 + 5224283/80-200 UGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGGUAAG-GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGA ...(((..............(((....)))(((((((......))))))).(((((((((.........)))))))-))........(((((..(((..........))).))))).))) ( -38.60) >Banth.0 31045 119 + 5227293/80-200 UGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCAUACGUAAUGGUAUGGUAUCCUUAUCUGAAUACAUAGGAUAAG-GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCUGGAGGA ...(((..............(((....)))(((((((......))))))).(((((((((.........)))))))-))........(((((..(((..........))).))))).))) ( -39.20) >consensus UGAUCCGAAGAUUUCCGAAUGGGGAAACCCACCACACGUAAUGGAAUGGUAUCCAUAUCUGAAUACAUAGGAUAAG_GAAGACAGACCCAGGGAACUGAAACAUCUAAGUACCCGGAGGA ...(((..............(((....)))(((((((......))))))).(((((((((.........))))))).))........(((((..(((..........))).))))).))) (-40.09 = -37.15 + -2.94)

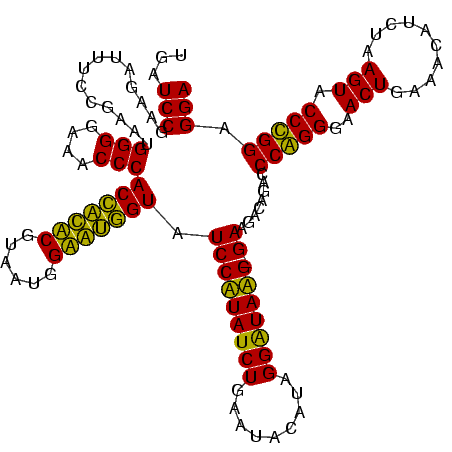
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 91.24 |
| Mean single sequence MFE | -37.05 |
| Consensus MFE | -36.22 |
| Energy contribution | -33.73 |
| Covariance contribution | -2.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.19 |
| Structure conservation index | 0.98 |
| SVM decision value | 3.25 |
| SVM RNA-class probability | 0.998836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/80-200 UCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACUGCUCCAUUACGAGCAGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCA (((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..(((((((((((......))))))))(((((((......)))))))))))))... ( -39.60) >Bsubt.0 32193 120 + 4214630/80-200 UCCUCCGGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUCUCAUAUCCUAUGAAUUCAGAUAUGGAUACCACUCCAUUACGAGUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCA (((.(((((.(((..((....)))))..)))))(((((..(((...........)))..)))))..(((((((((((......))))))))(((((((......)))))))))))))... ( -39.60) >Bcere.0 31024 119 + 5224283/80-200 UCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUC-CUUACCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCA .....((((.(((..((....)))))..))))((((((...((-((((..((........)).))))))((((((((......))))))))(((((((......)))))))..)))))). ( -32.70) >Banth.0 31045 119 + 5227293/80-200 UCCUCCAGGUACUUAGAUGUUUCAGUUCCCUGGGUCUGUCUUC-CUUAUCCUAUGUAUUCAGAUAAGGAUACCAUACCAUUACGUAUGGUGGGUUUCCCCAUUCGGAAAUCUUCGGAUCA .....((((.(((..((....)))))..))))((((((...((-((((((...........))))))))((((((((......))))))))(((((((......)))))))..)))))). ( -36.30) >consensus UCCUCCAGGUACUUAGAUGUUUCAGUUCCCCGGGUCUGCCUUC_CAUAUCCUAUGAAUUCAGAUAAGGAUACCACACCAUUACGAAUGGUGGGUUUCCCCAUUCGGAAAUCUCCGGAUCA .....((((.(((..((....)))))..))))((((((...((.((((((...((....))))))))))((((((((......))))))))(((((((......)))))))..)))))). (-36.22 = -33.73 + -2.50)

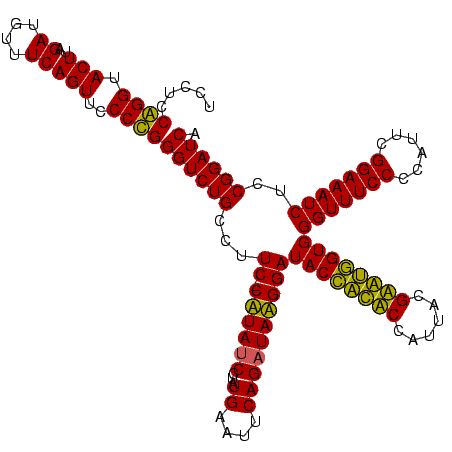
| Location | 36,508 – 36,627 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.96 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -34.40 |
| Energy contribution | -33.90 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.917092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 119 + 4222334/240-360 ACGGGA-ACAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUGUACGGAGUUACAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGC ..(((.-....)))...((((((((....)))))))).(((((...((.((((.(((...(((.......)))...))).(((..(.(((((....))))))..))).)))))).))))) ( -37.90) >Bsubt.0 32193 119 + 4214630/240-360 ACGGGA-UUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUGUACGGAGUUACAAAGGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGC ..(((.-....)))...((((((((....)))))))).(((((...((.((((.(((...(((.......)))...))).(((..(.(((((....))))))..))).)))))).))))) ( -37.90) >Bcere.0 31024 120 + 5224283/240-360 ACGGAACAUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAAC ..........((((...((((((((....))))))))...(.((.(((..(((((.....(((.......)))...)))))....(.(((((....))))))))).)).)))))...... ( -39.00) >Banth.0 31045 120 + 5227293/240-360 ACGGAACAUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAAC ..........((((...((((((((....))))))))...(.((.(((..(((((.....(((.......)))...)))))....(.(((((....))))))))).)).)))))...... ( -39.00) >consensus ACGGAA_AUAGCCCAAACCAAGAGGCUUGCCUCUUGGGGUUGUAGGACACUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGAGACCUGGAAAGGCCCGCCAUAGAAGGUAACAAC ..((........))...((((((((....)))))))).(((((...((..(((((.((...((((...........)))).))..(.(((((....))))))...)))))..)).))))) (-34.40 = -33.90 + -0.50)

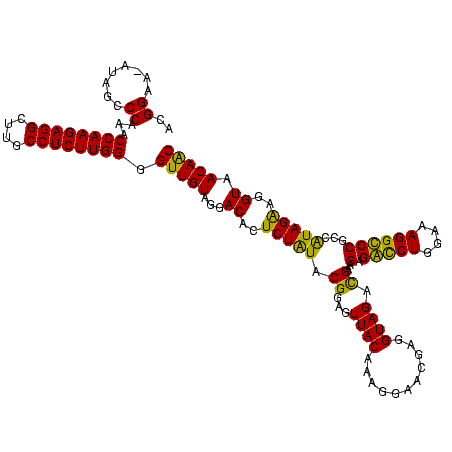
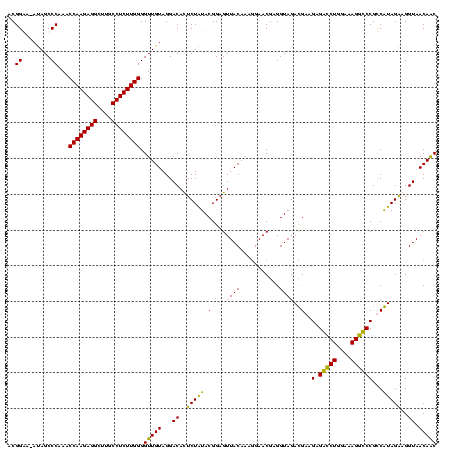
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -40.86 |
| Consensus MFE | -39.45 |
| Energy contribution | -38.76 |
| Covariance contribution | -0.69 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.48 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.995710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/280-400 UGUAGGACACUCUGUACGGAGUUACAAAAGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGCCCUGUAGUCAAAACUUCGUUCUCUCCUGAGUGGAUCCUGA ..((((((((((.....((((.......((((((((((...(((.(.(((((....))))))...(((((..........)))))..)))..)))))))))))))).)))))..))))). ( -39.61) >Bsubt.0 32193 120 + 4214630/280-400 UGUAGGACACUCUGUACGGAGUUACAAAGGAACGAGGUAGAUGAAGAGGUCUGGAAAGGCCCGCCAUAGGAGGUAACAGCCCUGUAGUCAAAACUUCGUUCUCUCCUGAGUGGAUCCUGA ..((((((((((.....((((.......((((((((((...(((.(.(((((....))))))...(((((..........)))))..)))..)))))))))))))).)))))..))))). ( -39.61) >Bcere.0 31024 120 + 5224283/280-400 UGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCCCGUAGUCGAAACUUCGUUCUCUCUUGAAUGUAUCCUGA ..((((((((((.....((((.......((((((((((.(((((.(.(((((....)))))).))...(.((((....)))))...)))...)))))))))))))).)))))..))))). ( -42.11) >Banth.0 31045 120 + 5227293/280-400 UGUAGGACAUUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGCGACCUGGAAAGGUCCGUCGUAGAGGGUAACAACCCCGUAGUCGAAACUUCGUUCUCUCUUGAAUGUAUCCUGA ..((((((((((.....((((.......((((((((((.(((((.(.(((((....)))))).))...(.((((....)))))...)))...)))))))))))))).)))))..))))). ( -42.11) >consensus UGUAGGACACUCUAUACGGAGUUACAAAGGAACGAGGUAGACGAAGAGACCUGGAAAGGCCCGCCAUAGAAGGUAACAACCCCGUAGUCAAAACUUCGUUCUCUCCUGAAUGGAUCCUGA ..((((((((((.....((((.......((((((((((..((((.(.(((((....)))))).))))...(((.......))).........)))))))))))))).)))))..))))). (-39.45 = -38.76 + -0.69)

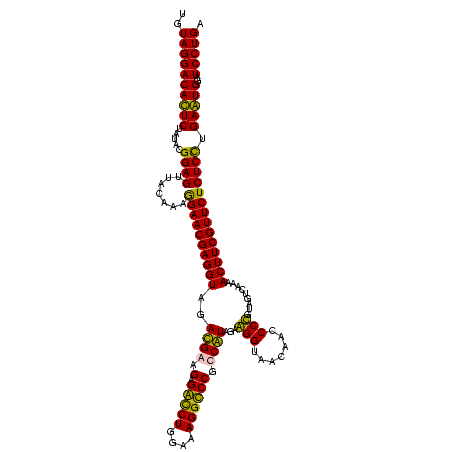
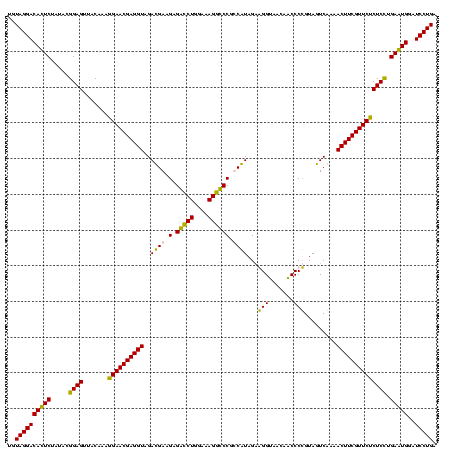
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.14 |
| Mean single sequence MFE | -33.60 |
| Consensus MFE | -31.81 |
| Energy contribution | -29.38 |
| Covariance contribution | -2.44 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.853826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/280-400 UCAGGAUCCACUCAGGAGAGAACGAAGUUUUGACUACAGGGCUGUUACCUCCUAUGGCGGGCCUUUCCAGACCUCUUCAUCUACCUCGUUCUUUUGUAACUCCGUACAGAGUGUCCUACA ..((((..(((((..((((((((((.((..(((....(((.((((((.......)))))).)))....((....)))))...)).))))))))))(((......))).)))))))))... ( -33.90) >Bsubt.0 32193 120 + 4214630/280-400 UCAGGAUCCACUCAGGAGAGAACGAAGUUUUGACUACAGGGCUGUUACCUCCUAUGGCGGGCCUUUCCAGACCUCUUCAUCUACCUCGUUCCUUUGUAACUCCGUACAGAGUGUCCUACA ..((((..(((((.((((.((((((.((..(((....(((.((((((.......)))))).)))....((....)))))...)).))))))))))(((......))).)))))))))... ( -31.90) >Bcere.0 31024 120 + 5224283/280-400 UCAGGAUACAUUCAAGAGAGAACGAAGUUUCGACUACGGGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUACA ..((((..(((((.((((.((((((.((...(((...(((((....)))))....((.((((((....))))).).))))).)).))))))))))(((......))).)))))))))... ( -34.30) >Banth.0 31045 120 + 5227293/280-400 UCAGGAUACAUUCAAGAGAGAACGAAGUUUCGACUACGGGGUUGUUACCCUCUACGACGGACCUUUCCAGGUCGCUUCGUCUACCUCGUUCCUUUGUAACUCCGUAUAGAAUGUCCUACA ..((((..(((((.((((.((((((.((...(((...(((((....)))))....((.((((((....))))).).))))).)).))))))))))(((......))).)))))))))... ( -34.30) >consensus UCAGGAUACACUCAAGAGAGAACGAAGUUUCGACUACAGGGCUGUUACCCCCUACGACGGACCUUUCCAGACCGCUUCAUCUACCUCGUUCCUUUGUAACUCCGUACAGAAUGUCCUACA ..((((..(((((.((((.((((((.((..(((.......(((((........))))).(((((....)))))...)))...)).))))))))))(((......))).)))))))))... (-31.81 = -29.38 + -2.44)

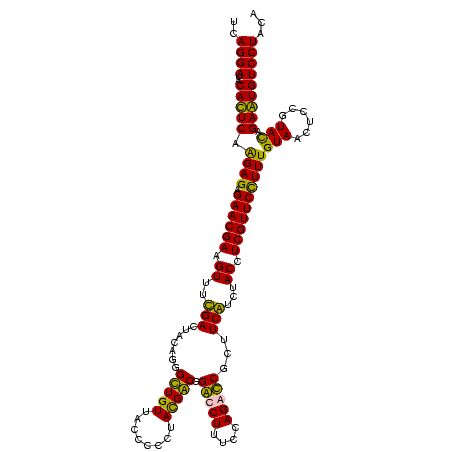
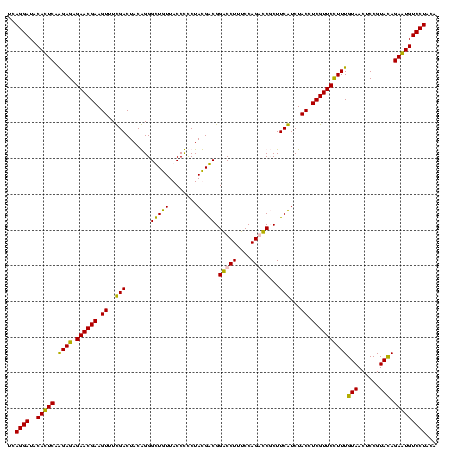
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 99.44 |
| Mean single sequence MFE | -42.20 |
| Consensus MFE | -41.87 |
| Energy contribution | -41.62 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.912534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/480-600 GAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGA ..(((....((....))....))).....(((((((....)))).((...((.....))...))....)))(((((((((((((..(((((....)))))..)))).)))...)))))). ( -42.40) >Bsubt.0 32193 120 + 4214630/480-600 GAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAGUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGA ..(((....((....))....))).....(((((((....)))).((...((.....))...))....)))(((((((((((((..(((((....)))))..)))).)))...)))))). ( -42.40) >Bcere.0 31024 120 + 5224283/480-600 GAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGA ..(((....((....))....))).....(((((((....)))).((...((.....))...))....)))(((((((..((((..(((((....)))))..))))......))))))). ( -42.00) >Banth.0 31045 120 + 5227293/480-600 GAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGA ..(((....((....))....))).....(((((((....)))).((...((.....))...))....)))(((((((..((((..(((((....)))))..))))......))))))). ( -42.00) >consensus GAACCAGUACCGUGAGGGAAAGGUGAAAAGCACCCCGGAAGGGGAGUGAAAGAGAUCCUGAAACCGUGUGCCUACAAAUAGUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGA ......((((((...((((..........((.((((....)))).))........)))).....)).))))(((((((..((((..(((((....)))))..))))......))))))). (-41.87 = -41.62 + -0.25)

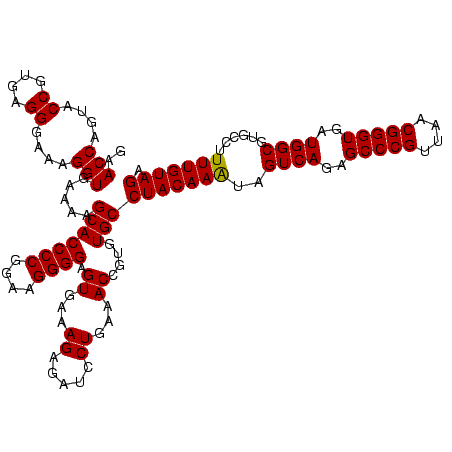
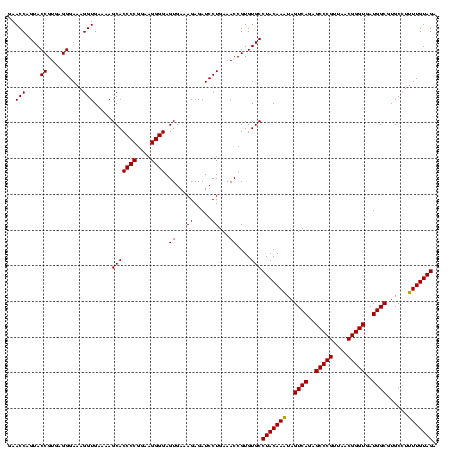
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.93 |
| Mean single sequence MFE | -44.75 |
| Consensus MFE | -44.30 |
| Energy contribution | -43.55 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.98 |
| SVM RNA-class probability | 0.894849 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/560-680 GUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGUUUGAAAAGACGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG ((((..(((((....)))))..))))..(((((...((((......(((.((.......)))))(((..((((..((((....))))..)))))))((....))...))))...))))). ( -45.20) >Bsubt.0 32193 119 + 4214630/560-680 GUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGCA-GAAGAUGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG ((((..(((((....)))))..))))..(((((...((((......(((.((.......)))))(((..((((..(((-.....)))..)))))))((....))...))))...))))). ( -44.80) >Bcere.0 31024 119 + 5224283/560-680 GUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG ((((..(((((....)))))..))))..(((((...((((......(((.((.......)))))(((..((((..(((-.....)))..)))))))((....))...))))...))))). ( -44.50) >Banth.0 31045 119 + 5227293/560-680 GUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG ((((..(((((....)))))..))))..(((((...((((......(((.((.......)))))(((..((((..(((-.....)))..)))))))((....))...))))...))))). ( -44.50) >consensus GUCAGAGCCCGUUAACGGGUGAUGGCGUGCCUUUUGUAGAAUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGCU_GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCG ((((..(((((....)))))..))))..(((((...((((......(((.((.......)))))(((..((((..(((......)))..)))))))((....))...))))...))))). (-44.30 = -43.55 + -0.75)


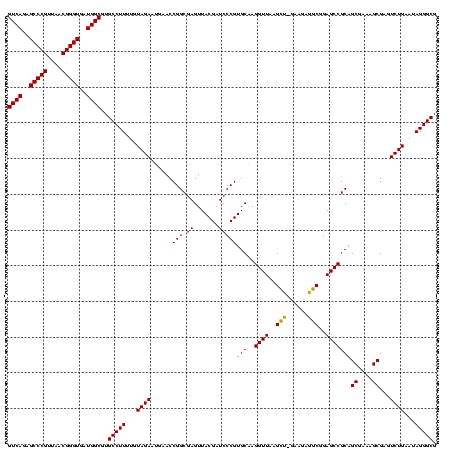
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.25 |
| Mean single sequence MFE | -40.95 |
| Consensus MFE | -40.75 |
| Energy contribution | -39.75 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.44 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/600-720 AUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGUUUGAAAAGACGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCAUGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUAC ....(((((((.(((((((((.(((((..((((..((((....))))..))))((.((....))..((((.....))))))....))))).)))))).))).))...)).)))....... ( -42.90) >Bsubt.0 32193 119 + 4214630/600-720 AUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGCA-GAAGAUGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCAUGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUAC ....(((((((.(((((((((.(((((..((((..(((-.....)))..))))((.((....))..((((.....))))))....))))).)))))).))).))...)).)))....... ( -42.50) >Bcere.0 31024 118 + 5224283/600-720 AUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGUUU-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUAC ....(((((((.(((((((((.(((((..((((..(((-.....)))..))))((.((....))..((((.....))))))..-.))))).)))))).))).))...)).)))....... ( -39.20) >Banth.0 31045 118 + 5227293/600-720 AUGAACCGGCGAGUUACGAUCCCGUGCGAGGUUAAGCU-GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGUUU-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUAC ....(((((((.(((((((((.(((((..((((..(((-.....)))..))))((.((....))..((((.....))))))..-.))))).)))))).))).))...)).)))....... ( -39.20) >consensus AUGAACCGGCGAGUUACGAUCCCGUGCAAGGUUAAGCU_GAAGAGGCGGAGCCGCAGCGAAAGCGAGUCUGAAUAGGGCGCAU_AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUAC ....(((((((.(((((((((.(((((..((((..(((......)))..))))((.((....))..((((.....))))))....))))).)))))).))).))...)).)))....... (-40.75 = -39.75 + -1.00)

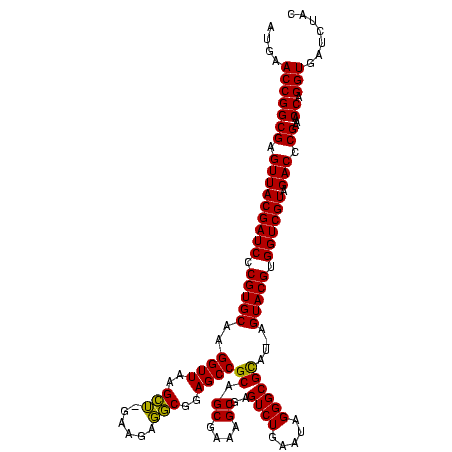
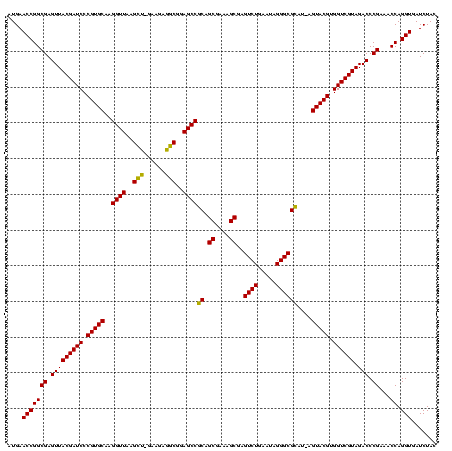
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -42.50 |
| Consensus MFE | -42.50 |
| Energy contribution | -42.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.63 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/680-800 CAUGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUG .......((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).... ( -42.50) >Bsubt.0 32193 120 + 4214630/680-800 CAUGAGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUG .......((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).... ( -42.50) >Bcere.0 31024 119 + 5224283/680-800 UUU-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUG ...-...((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).... ( -42.50) >Banth.0 31045 119 + 5227293/680-800 UUU-AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUG ...-...((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).... ( -42.50) >consensus CAU_AGUACGUGGUCGUAGACCCGAAACCAGGUGAUCUACCCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUG .......((((((..((((((((.......)).).)))))))))))((.((((...((((((......))))))....))))...(((.(((((........))))))))....)).... (-42.50 = -42.50 + 0.00)

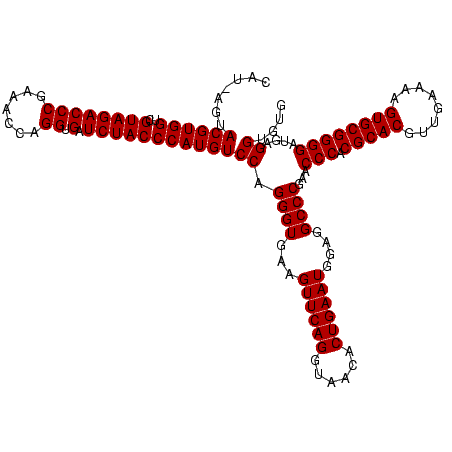
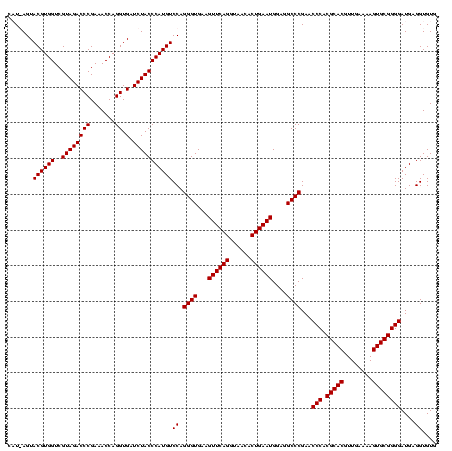
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -40.75 |
| Energy contribution | -40.25 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.96 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.861370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/720-840 CCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGU ((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))).((((..(((........)))..))))............ ( -42.70) >Bsubt.0 32193 120 + 4214630/720-840 CCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGU ((((..(((((((...((((((......))))))....))))...(((.(((((........)))))))).)).)..)))).((((..(((........)))..))))............ ( -42.70) >Bcere.0 31024 120 + 5224283/720-840 CCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGU ..((.((((((.(((.((((((......))))))((((.((((.((((((((.(((........)))........))))))))..))).)...))))..)))..)))))).))....... ( -42.40) >Banth.0 31045 120 + 5227293/720-840 CCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGU ..((.((((((.(((.((((((......))))))((((.((((.((((((((.(((........)))........))))))))..))).)...))))..)))..)))))).))....... ( -42.40) >consensus CCAUGUCCAGGGUGAAGUUCAGGUAACACUGAAUGGAGGCCCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUGCCAAUCGAACCUGGAGAUAGCUGGU ..((.((((((.....((((((......)))))).((.(((((.((((((((((........)))))....)).))).)))))...(((.....)))..))...)))))).))....... (-40.75 = -40.25 + -0.50)

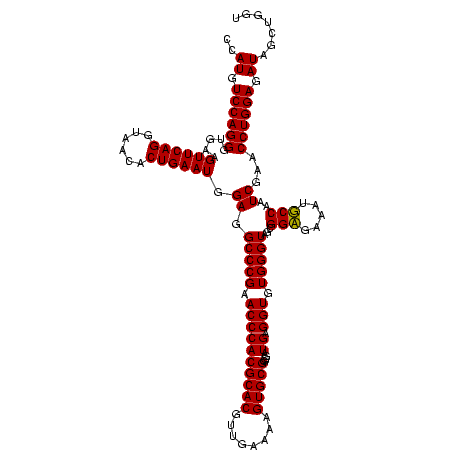
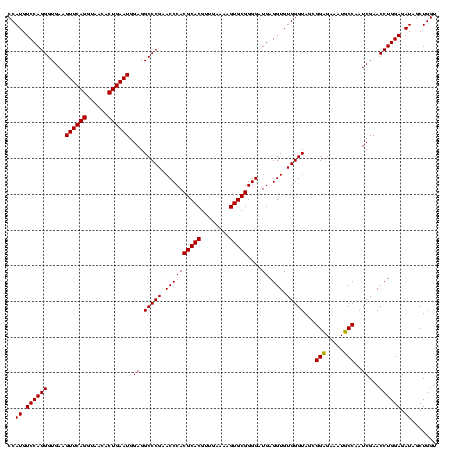
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -34.45 |
| Consensus MFE | -32.35 |
| Energy contribution | -32.35 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.902968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/720-840 ACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGG .....((((.((((((......(((.......(((..((((((...........((((........))))))))))..)))...)))(((((......)))))......)))))).)))) ( -33.60) >Bsubt.0 32193 120 + 4214630/720-840 ACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGG .....((((.((((((......(((.......(((..((((((...........((((........))))))))))..)))...)))(((((......)))))......)))))).)))) ( -33.60) >Bcere.0 31024 120 + 5224283/720-840 ACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGG .....((((.((((((......((((.....((((..((((((...........((((........)))))))))).))))..))))(((((......)))))......)))))).)))) ( -35.30) >Banth.0 31045 120 + 5227293/720-840 ACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGG .....((((.((((((......((((.....((((..((((((...........((((........)))))))))).))))..))))(((((......)))))......)))))).)))) ( -35.30) >consensus ACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCACCCCUACCCACACCUCAUCCCCGCACUUUUCAACGUGCGUGGGUUCGGGCCUCCAUUCAGUGUUACCUGAACUUCACCCUGGACAUGG .....((((.(((((((((((..((.......))................((((((((........))))).))).)))))))....(((((......)))))........)))).)))) (-32.35 = -32.35 + 0.00)



| Location | 36,508 – 36,627 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.66 |
| Mean single sequence MFE | -39.90 |
| Consensus MFE | -39.40 |
| Energy contribution | -38.40 |
| Covariance contribution | -1.00 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.99 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.598378 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 119 + 4222334/760-880 CCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAG-GUAAGA ((...(((.(((((........)))))))).((((((.((((((((..(((........)))..))))((((........))))))))...(((((....))))))))))).)-)..... ( -41.40) >Bsubt.0 32193 119 + 4214630/760-880 CCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGGGGUGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCUCCGAAAUAGCUUUAGGGCUAGCCUCAAG-GUAAGA ((...(((.(((((........)))))))).((((((.((((((((..(((........)))..))))((((........))))))))...(((((....))))))))))).)-)..... ( -41.40) >Bcere.0 31024 120 + 5224283/760-880 CCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAAGA .....(((.(((((........)))))))).((((((.((((....((((...))))....(((((.((......)))))))..))))...(((((....)))))))))))......... ( -38.40) >Banth.0 31045 120 + 5227293/760-880 CCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUUCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUUAAGUGUAAGA .....(((.(((((........)))))))).((((((.((((....((((...))))....(((((.((......)))))))..))))...(((((....)))))))))))......... ( -38.40) >consensus CCGAACCCACGCACGUUGAAAAGUGCGGGGAUGAGGUGUGGGUAGCGGAGAAAUGCCAAUCGAACCUGGAGAUAGCUGGUUCUCCCCGAAAUAGCUUUAGGGCUAGCCUCAAG_GUAAGA .....(((.(((((........)))))))).((((((.((((....(((.....)))....(((((.((......)))))))..))))...(((((....)))))))))))......... (-39.40 = -38.40 + -1.00)

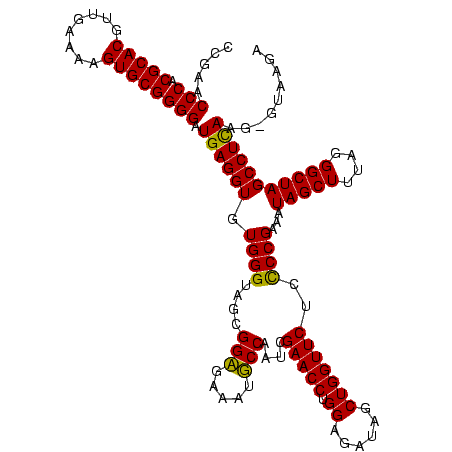
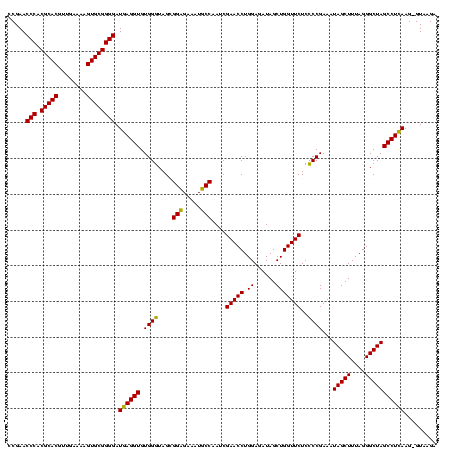
| Location | 36,508 – 36,627 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -33.15 |
| Consensus MFE | -26.10 |
| Energy contribution | -27.35 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588198 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 119 + 4222334/800-920 GUAGGGGCCCCUAGUCCAAUCAGUGCUCUACCUCCAAGACUCUUAC-CUUGAGGCUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACC (((((((.....(((((((((....((((.((((.(((........-))))))).((((......))))....)))).(((((...........))))))))))).)))...))))))). ( -38.70) >Bsubt.0 32193 119 + 4214630/800-920 GUAGGGGCCCCUAGUCCAAUCAGUGCUCUACCUCCAAGACUCUUAC-CUUGAGGCUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGCAUUUCACCCCUACC (((((((.....(((((((((....((((.((((.(((........-))))))).((((......))))....)))).(((((...........))))))))))).)))...))))))). ( -38.70) >Bcere.0 31024 120 + 5224283/800-920 AUGAGGACCCCUAGUCCAAUCAGUGCUCUACCUCCAAGACUCUUACACUUAAGGCUAGCCCUAAAGCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACC ....(((.......(((((((..........((((.....(((((....))))).((((......))))....)))).(((((...........)))))))))))).....)))...... ( -27.60) >Banth.0 31045 120 + 5227293/800-920 AUGAGGACCCCUAGUCCAAUCAGUGCUCUACCUCCAAGACUCUUACACUUAAGGCUAGCCCUAAAGCUAUUUCGGGGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCUCCGCUACC ....(((.......(((((((..........((((.....(((((....))))).((((......))))....)))).(((((...........)))))))))))).....)))...... ( -27.60) >consensus AUAAGGACCCCUAGUCCAAUCAGUGCUCUACCUCCAAGACUCUUAC_CUUAAGGCUAGCCCUAAAGCUAUUUCGGAGAGAACCAGCUAUCUCCAGGUUCGAUUGGAAUUUCACCCCUACC (((((((.....(((((((((..........((((.....(((((....))))).((((......))))....)))).(((((...........))))))))))).)))...))))))). (-26.10 = -27.35 + 1.25)

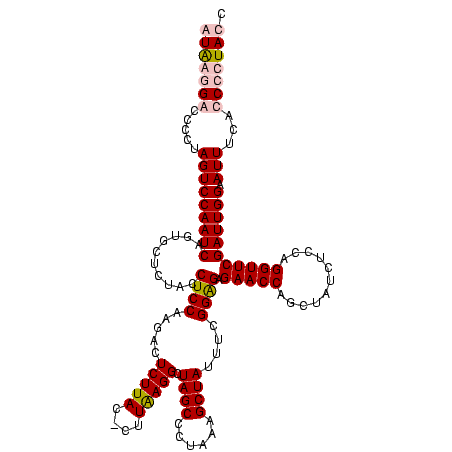
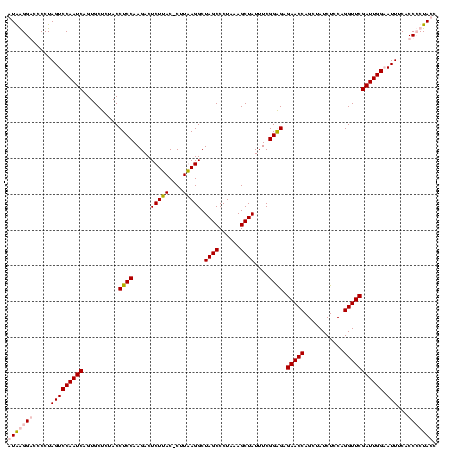
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.67 |
| Mean single sequence MFE | -37.65 |
| Consensus MFE | -37.30 |
| Energy contribution | -36.80 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.99 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978866 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/1040-1160 GUAUACGUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAU ...(((.....))).....((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..))))...... ( -38.20) >Bsubt.0 32193 120 + 4214630/1040-1160 GUAUACGUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUGGUCGAGUGACUCUGCGCCGAAAAU ...(((.....))).....((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..))))...... ( -38.20) >Bcere.0 31024 120 + 5224283/1040-1160 GUGUGUAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAU ...................((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..))))...... ( -37.10) >Banth.0 31045 120 + 5227293/1040-1160 GUGUGUAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACUAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAU ...................((.((..((((..(((.(((.....(((((.(((((.......))))).)))))....((((.((....)).)))).))))))..))))..))))...... ( -37.10) >consensus GUAUACAUUAAGUGGAAAAGGAUGUGGAGUUGCUUAGACAACCAGGAUGUUGGCUUAGAAGCAGCCACCAUUUAAAGAGUGCGUAAUAGCUCACUAGUCGAGUGACUCUGCGCCGAAAAU ...................((.((..((((..(((.....(((((((((.(((((.......))))).))))....((((........)))).))))).)))..))))..))))...... (-37.30 = -36.80 + -0.50)

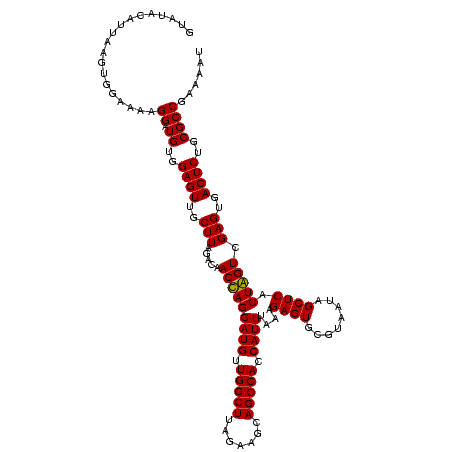
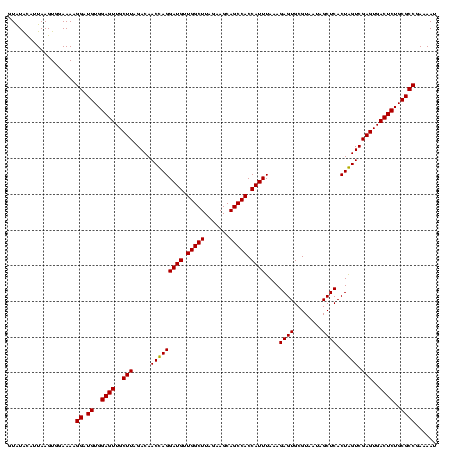
| Location | 36,508 – 36,625 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -40.42 |
| Consensus MFE | -40.22 |
| Energy contribution | -38.35 |
| Covariance contribution | -1.87 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.88 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975199 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 117 + 4222334/1160-1280 GUACCGGGGCUAAACGUAUCACCGAAGCUGCGGACUGUUCU---UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAG ...(((.((((...((......)).)))).)))(((((((.---...))))))).........((.(((((((.(((.(...(((((.((....)).)))))).))).)))))))))... ( -44.70) >Bsubt.0 32193 116 + 4214630/1160-1280 GUACCGGGGCUAAACGUAUCACCGAAGCUGCGGACUGUUCU---U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAG ((.(((.((((...((......)).)))).))))).(((((---(-(..((....)).))))))).(((((((.(((.(...(((((.((....)).)))))).))).)))))))..... ( -44.90) >Bcere.0 31024 120 + 5224283/1160-1280 GUACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAG ...(((.((((..............)))).)))(((((((((...))))))))).........((.(((((((.((......(((((.((....)).)))))...)).)))))))))... ( -36.04) >Banth.0 31045 120 + 5227293/1160-1280 GUACCGGGGCUAAAUACACCACCGAAGCUGCGGAUUGAUACCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAG ...(((.((((..............)))).)))(((((((((...))))))))).........((.(((((((.((......(((((.((....)).)))))...)).)))))))))... ( -36.04) >consensus GUACCGGGGCUAAACACACCACCGAAGCUGCGGACUGAUAC___UGCGAACAGUGGUAGGAGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAG .(((....(((...(.((((.(((......)))((((((((.....)))))))))))).)..))).(((((((.((......(((((.((....)).)))))...)).)))))))))).. (-40.22 = -38.35 + -1.87)

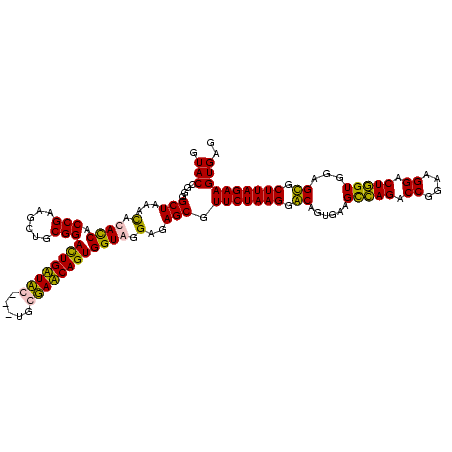
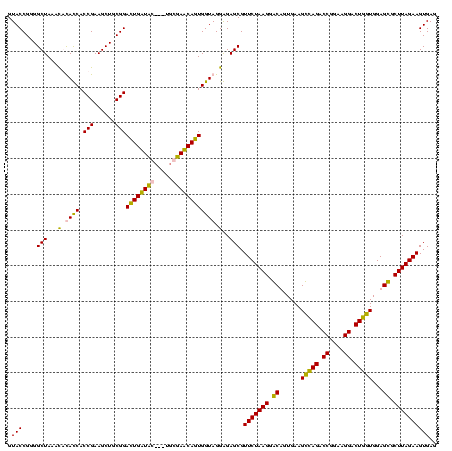
| Location | 36,508 – 36,625 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.15 |
| Mean single sequence MFE | -31.23 |
| Consensus MFE | -26.23 |
| Energy contribution | -25.85 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.944350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 117 + 4222334/1160-1280 CUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCGCA---AGAACAGUCCGCAGCUUCGGUGAUACGUUUAGCCCCGGUAC .....(((((((.(((...((((.((....)).))))......))).)))))))............(((((((...---.)))))))(((..(((.((......))...)))..)))... ( -36.60) >Bsubt.0 32193 116 + 4214630/1160-1280 CUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCG-A---AGAACAGUCCGCAGCUUCGGUGAUACGUUUAGCCCCGGUAC .....(((((((.(((...((((.((....)).))))......))).)))))))............(((((((.-.---.)))))))(((..(((.((......))...)))..)))... ( -31.90) >Bcere.0 31024 120 + 5224283/1160-1280 CUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGUAC .....(((((((.((.....(((.((....)).)))........)).)))))))........((((..(((((((...)))))))............((.(((.......))))))))). ( -28.22) >Banth.0 31045 120 + 5227293/1160-1280 CUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGGUAUCAAUCCGCAGCUUCGGUGGUGUAUUUAGCCCCGGUAC .....(((((((.((.....(((.((....)).)))........)).)))))))........((((..(((((((...)))))))............((.(((.......))))))))). ( -28.22) >consensus CUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACAGCCCUUAGAACGCUCCCCUACCACUGAUACCA___AGAACAAUCCGCAGCUUCGGUGAUACAUUUAGCCCCGGUAC .....(((((((.((....((((.((....)).)))).......)).)))))))........((((((((((((.....)))))))).....((....))...............)))). (-26.23 = -25.85 + -0.38)

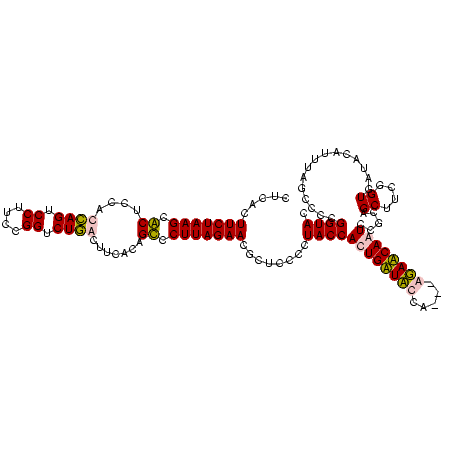
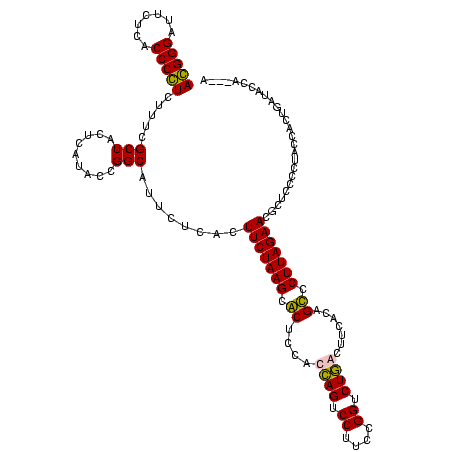
| Location | 36,508 – 36,625 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -38.05 |
| Consensus MFE | -37.33 |
| Energy contribution | -35.70 |
| Covariance contribution | -1.63 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.98 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997005 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 117 + 4222334/1200-1320 U---UGCGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCGAGACCGGAAGGACUCGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGGGGUGAGAAUCCCCU (---(((...((.(((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))))...))....))))...(((((.......))))) ( -43.30) >Bsubt.0 32193 116 + 4214630/1200-1320 U---U-CGAACAGUGGUAGGAGAGCGUUCUAAGGGCUGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGAGGGGUGAGAAUCCCCU (---(-((.((...((((.....((.(((((((.(((.(...(((((.((....)).)))))).))).))))))))).....)))).)).))))...(....)(((((.......))))) ( -42.90) >Bcere.0 31024 120 + 5224283/1200-1320 CCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCGU ....((.(((((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))...))).)).))....(((((.......))))) ( -33.00) >Banth.0 31045 120 + 5227293/1200-1320 CCAAUGGUAUCAGUGGUAGGGGAGCGUUCUAAGGACAGUGAAGUCAGACCGGAAGGACUGGUGGAGUGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCGU ....((.(((((.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....)))))...))).)).))....(((((.......))))) ( -33.00) >consensus C___UGCGAACAGUGGUAGGAGAGCGUUCUAAGGACAGUGAAGCCAGACCGGAAGGACUGGUGGAGCGCUUAGAAGUGAGAAUGCCGGUAUGAGUAGCGAAAGACGGGUGAGAAUCCCCU ...........(.(((((.....((.(((((((.((......(((((.((....)).)))))...)).))))))))).....))))).)........(....)(((((.......))))) (-37.33 = -35.70 + -1.63)


| Location | 36,508 – 36,625 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.93 |
| Mean single sequence MFE | -23.36 |
| Consensus MFE | -21.77 |
| Energy contribution | -20.90 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.833889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 117 + 4222334/1200-1320 AGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACGAGUCCUUCCGGUCUCGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCGCA---A ((((((................(((.........)))........(((((((.(((...((((.((....)).))))......))).)))))))...)))))).............---. ( -25.50) >Bsubt.0 32193 116 + 4214630/1200-1320 AGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCGCUCCACCAGUCCUUCCGGUCUGGCUUCACAGCCCUUAGAACGCUCUCCUACCACUGUUCG-A---A ((((((................(((.........)))........(((((((.(((...((((.((....)).))))......))).)))))))...))))))...........-.---. ( -26.10) >Bcere.0 31024 120 + 5224283/1200-1320 ACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGG (((((.......))))).....(((.........)))........(((((((.((.....(((.((....)).)))........)).)))))))..........(((.((....)).))) ( -20.92) >Banth.0 31045 120 + 5227293/1200-1320 ACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACUGUCCUUAGAACGCUCCCCUACCACUGAUACCAUUGG (((((.......))))).....(((.........)))........(((((((.((.....(((.((....)).)))........)).)))))))..........(((.((....)).))) ( -20.92) >consensus ACGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUUCUCACUUCUAAGCACUCCACCAGUCCUUCCGGUCUGACUUCACAGCCCUUAGAACGCUCCCCUACCACUGAUACCA___A (((((.......))))).....(((.........)))........(((((((.((....((((.((....)).)))).......)).))))))).......................... (-21.77 = -20.90 + -0.87)

| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.78 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -41.00 |
| Energy contribution | -40.50 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.61 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.634158 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/1280-1400 UACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUU ...(((....((((((.(((((((...........))))))).)).).)))..(((.(((...))).)))....(((.(((((((.......))))))).)))...........)))... ( -40.90) >Bsubt.0 32193 120 + 4214630/1280-1400 UACGCCUUUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGAGGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUU ...(((....((((((.(((((((...........))))))).)).).)))..(((.(((...))).)))....(((.(((((((.......))))))).)))...........)))... ( -40.90) >Bcere.0 31024 120 + 5224283/1280-1400 UACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUU ...(((((((((((((.(((((((...........))))))).)).).)))((((...))))......))))..(((.(((((((.......))))))).)))...........)))... ( -41.50) >Banth.0 31045 120 + 5227293/1280-1400 UACGCCUGUCGGCCUCGGCUUAGGUCCUGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCGUCUUUCGCUACUCAUACCGGCAUU ...(((((((((((((.(((((((...........))))))).)).).)))((((...))))......))))..(((.(((((((.......))))))).)))...........)))... ( -41.50) >consensus UACGCCUGUCGGCCUCGGCUUAGGUCCCGACUAACCCUGAGCGGACGAGCCUUCCUCAGGAAACCUUAGGCAUUCGGUGGACGGGAUUCUCACCCCUCUUUCGCUACUCAUACCGGCAUU ...(((....((((((.(((((((...........))))))).)).).)))..(((.(((...))).)))....(((.(((((((.......))))))).)))...........)))... (-41.00 = -40.50 + -0.50)

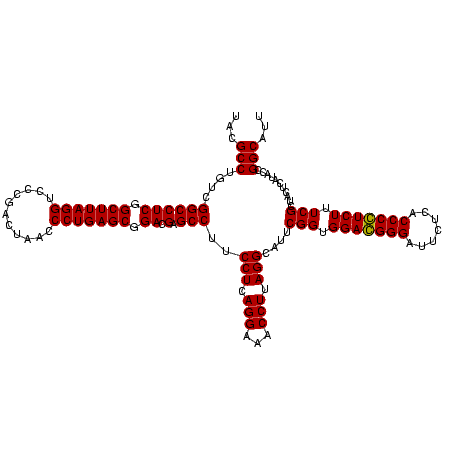
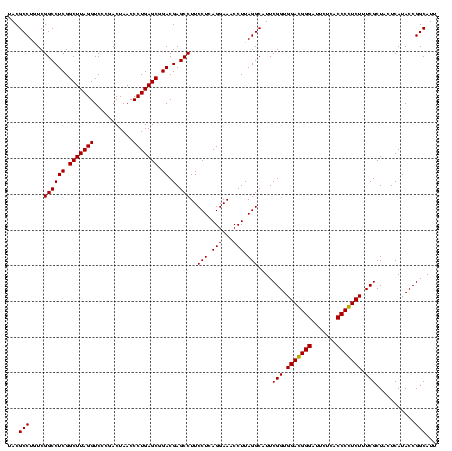
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -29.51 |
| Energy contribution | -28.01 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.606823 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/1400-1520 UCCCAGCCUAACUGCUUGGACGCGGAUAUCCAAUACCGCGCUUGCCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAAUGGUGAGGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCC (((.(((......))).)))(((((..........)))))................((.(((.((((((.....))))).).)))(((((.(((((........)))))...))))))). ( -36.10) >Bsubt.0 32193 120 + 4214630/1400-1520 UCCCAGCCUAACUGCUUGGACGCGGAUAUCCAAUACCGCGCUUACCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAAUGGUGAGGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCC (((.(((......))).)))(((((..........)))))................((.(((.((((((.....))))).).)))(((((.(((((........)))))...))))))). ( -36.10) >Bcere.0 31024 120 + 5224283/1400-1520 UAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCC .............((.(((((((((..........))))(((((((.((((.....(((.........)))......)))))))))))...(((((........))))).)))))..)). ( -28.20) >Banth.0 31045 120 + 5227293/1400-1520 UAUCAGCCUAACUGCUUGGACGUGCACAACCAAUCGCACGCUUCUUCUAUCCUUCUGCGUCCCUCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCC .............((.(((((((((..........))))(((((((.((((.....(((.........)))......)))))))))))...(((((........))))).)))))..)). ( -28.20) >consensus UACCAGCCUAACUGCUUGGACGCGCACAACCAAUACCACGCUUCCCCUAUCCUCCUGCGUCCCCCCAUUGCUCAAACGAUAAAGAGGUGGUACAGGAAUAUCAACCUGUUGUCCAUCGCC ....(((......))).((((((((..........))))((...............))))))((.(((((......))))).)).(((((.(((((........)))))...)))))... (-29.51 = -28.01 + -1.50)

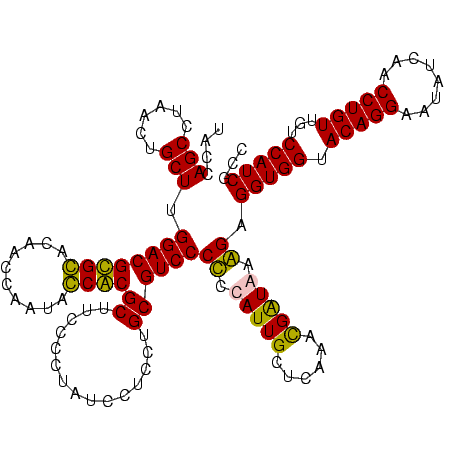
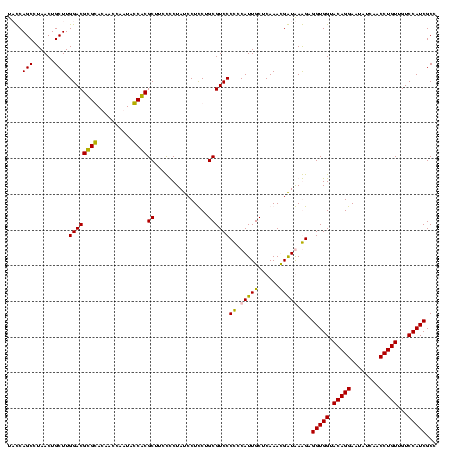
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.84 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -29.33 |
| Energy contribution | -32.95 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.600004 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/1720-1840 CGUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUA ....(((((.(((((..(.(.(((((.((.((((....)))))).)))))).).(((.....((.(((....)))....)).....)))...))))).....(((....))))))))... ( -42.10) >Bsubt.0 32193 120 + 4214630/1720-1840 CGUAACUUCGGGAGAAGGGGUGCUCUGUUAGGGUGCAAGCCCGAGAGAGCCGCAGUGAAUAGGCCCAGGCGACUGUUUAGCAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUA ....(((((.(((((..(.(.(((((.((.((((....)))))).)))))).).(((.....((.(((....)))....)).....)))...))))).....(((....))))))))... ( -42.10) >Bcere.0 31024 109 + 5224283/1720-1840 CGUAACUUCGGGAGAAGGGGUGCUUUCUUAACG-----------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUA ....(((((.(((((..(((((((((((....)-----------))))))............))))..((...(((((.....)))))..))))))).....(((....))))))))... ( -33.50) >Banth.0 31045 109 + 5227293/1720-1840 CGUAACUUCGGGAGAAGGGGUGCUUUCUUAACG-----------GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUA ....(((((.(((((..(((((((((((....)-----------))))))............))))..((...(((((.....)))))..))))))).....(((....))))))))... ( -33.50) >consensus CGUAACUUCGGGAGAAGGGGUGCUCUCUUAACG___________GAAAGCCGCAGUGAAUAGGCCCAAGCGACUGUUUAGCAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUA ....(((((.(((((...(((((((((...((((....))))..)))))).((..(((((((((....))..)))))))))....)).)...))))).....(((....))))))))... (-29.33 = -32.95 + 3.62)

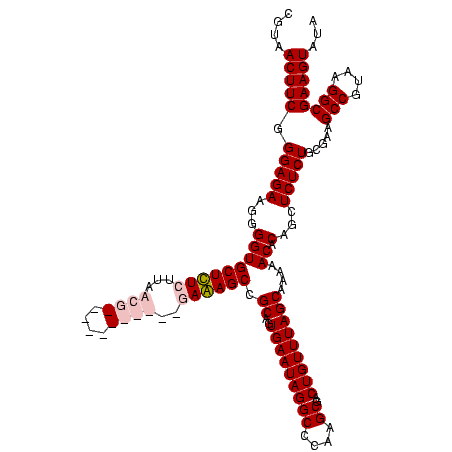
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -39.55 |
| Energy contribution | -41.05 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.980007 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/1800-1920 CAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAA .............((.(((...(((....)))...))).))((((........))))..((((((..((((..((...((((((.((....))..))))))...))..)))))))))).. ( -43.30) >Bsubt.0 32193 120 + 4214630/1800-1920 CAAAAACACAGGUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAA .............((.(((...(((....)))...))).))((((........))))..((((((..((((..((...((((((.((....))..))))))...))..)))))))))).. ( -43.30) >Bcere.0 31024 120 + 5224283/1800-1920 CAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAA ......(((..((((.(((...(((....)))...))).)))).))).((((.....))))((((..((((..((...((((((.((....))..))))))...))..)))))))).... ( -42.10) >Banth.0 31045 120 + 5227293/1800-1920 CAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAA ......(((..((((.(((...(((....)))...))).)))).))).((((.....))))((((..((((..((...((((((.((....))..))))))...))..)))))))).... ( -42.10) >consensus CAAAAACACAGCUCUCUGCGAAGCCGUAAGGCGAAGUAUAGGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAAGAGCGCUUAGCGUAAGCGAAGCUCCGAACUGAAGCCCCAGUAAA ..........(((((((((...(((....)))...))..)))))))..((((.....))))((((..((((..((...((((((.((....))..))))))...))..)))))))).... (-39.55 = -41.05 + 1.50)

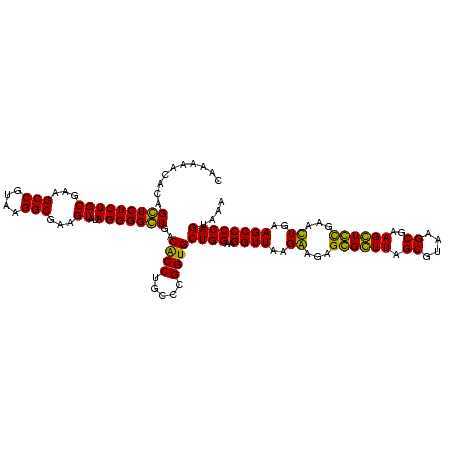
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.44 |
| Mean single sequence MFE | -43.70 |
| Consensus MFE | -42.35 |
| Energy contribution | -42.35 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.871525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/1840-1960 GGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUC ...(((...(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...)))))))....... ( -45.10) >Bsubt.0 32193 120 + 4214630/1840-1960 GGGGCUGACGCCUGCCCGGUGCUGGAAGGUUAAGAGGAGCGCUUAGCGUAAGCGAAGGUGCGAAUUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUC ...(((...(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...)))))))....... ( -45.10) >Bcere.0 31024 120 + 5224283/1840-1960 GGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUC ...(....)(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...)))).......... ( -42.30) >Banth.0 31045 120 + 5227293/1840-1960 GGGGGUGACACCUGCCCGGUGCUGGAAGGUUAAGGAGAGGGGUUAGCGUAAGCGAAGCUCUGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUC ...(....)(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...)))).......... ( -42.30) >consensus GGGGCUGACACCUGCCCGGUGCUGGAAGGUUAAGAAGAGCGCUUAGCGUAAGCGAAGCUCCGAACUGAAGCCCCAGUAAACGGCGGCCGUAACUAUAACGGUCCUAAGGUAGCGAAAUUC .....((..(((((((...((((((..((((..((...((((((.((....))..))))))...))..))))))))))...)))((((((.......))))))...))))..))...... (-42.35 = -42.35 + 0.00)

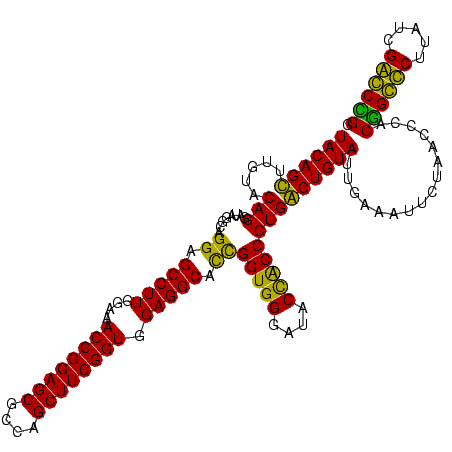
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -49.18 |
| Consensus MFE | -51.55 |
| Energy contribution | -48.42 |
| Covariance contribution | -3.12 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.08 |
| Structure conservation index | 1.05 |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.994547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2120-2240 UACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGG (((((((.(..............(((((((....((((((((....)))))))).))))).))(((((....)))))).)))))))................((((((.....)))))). ( -48.20) >Bsubt.0 32193 120 + 4214630/2120-2240 UACAGCUUGUACAGGAUAGGUAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGG (((((((.(..............(((((((....((((((((....)))))))).))))).))(((((....)))))).)))))))................((((((.....)))))). ( -49.70) >Bcere.0 31024 120 + 5224283/2120-2240 UACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGG .....((.((((((..(((((((..(((((....((((((((....)))))))).))))).)))((((....)))))))).)))))).))............((((((.....)))))). ( -49.40) >Banth.0 31045 120 + 5227293/2120-2240 UACAGUUUGUACAGGAUAGGCGGGAGCCUUUGAAACCGGAGCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGG .....((.((((((..(((((((..(((((....((((((((....)))))))).))))).)))((((....)))))))).)))))).))............((((((.....)))))). ( -49.40) >consensus UACAGCUUGUACAGGAUAGGCAGGAGCCUUGGAAACCGGAGCGCCAGCUUCGGUGGAGGCACCGGUGGGAUACCACCCUGACUGUAUUGAAAUUCUAACCCACGGCCCUUAUCGACCCGG ((((((.....(((........((.(((((....((((((((....)))))))).))))).))(((((....))))))))))))))................((((((.....)))))). (-51.55 = -48.42 + -3.12)

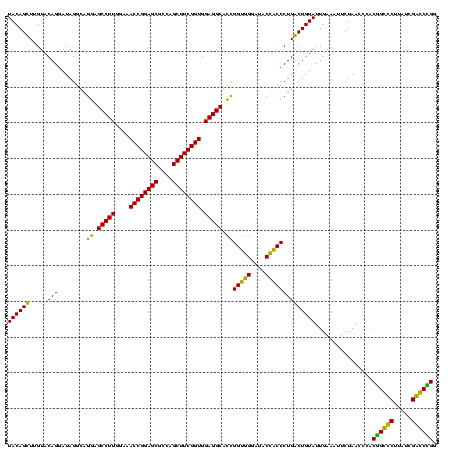
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.06 |
| Mean single sequence MFE | -40.23 |
| Consensus MFE | -38.94 |
| Energy contribution | -36.06 |
| Covariance contribution | -2.87 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.58 |
| SVM RNA-class probability | 0.790431 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2120-2240 CCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCUGUACAAGCUGUA .((((((.....))))))((((.(((.(((.......((((..((..(((((.....)))))..))...((((((((....))))))))...)))))))..))).))))........... ( -41.61) >Bsubt.0 32193 120 + 4214630/2120-2240 CCCGCCCGAUAAGGGCGGCGGGUUAGAAGGUCAAUACAGCCAGGGUAGUAUCCCACCGAUGCCUCCACCGAAGCUGGCGCUCCGGUUUCCAAGGCUCCUACCUAUCCUGUACAAGCUGUA .((((((.....))))))((((.(((.(((.......((((..((..(((((.....)))))..))...((((((((....))))))))...)))))))..))).))))........... ( -43.11) >Bcere.0 31024 120 + 5224283/2120-2240 CCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGUA .((((((.....))))))................(((((...(((((((.....)))...((((..((((.(((....))).)))).....))))....))))...)))))......... ( -38.10) >Banth.0 31045 120 + 5227293/2120-2240 CCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGUCAGGGCGGUAUCCCACCAGCGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCGCCUAUCCUGUACAAACUGUA .((((((.....))))))................(((((...(((((((.....)))...((((..((((.(((....))).)))).....))))....))))...)))))......... ( -38.10) >consensus CCGGCCCGAUAAGACCCGCAGGUUAGAAGGUCAAUACAGCCAGGGCAGUAUCCCACCAACGCCUCCACCGAAGCUAGCGCUCCGGUUUCAAAGGCUCCCACCUAUCCUGUACAAACUGUA .((((((.....))))))..((((.(.....)..(((((...((((((............((((..((((.(((....))).)))).....))))..))))))...)))))..))))... (-38.94 = -36.06 + -2.87)

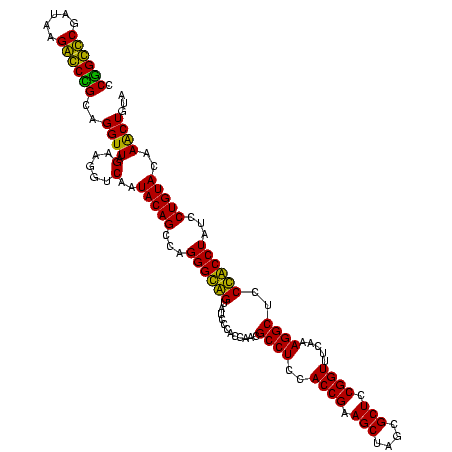
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.28 |
| Mean single sequence MFE | -52.12 |
| Consensus MFE | -50.50 |
| Energy contribution | -47.38 |
| Covariance contribution | -3.12 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2160-2280 GCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCC (((((.((((((((.(((.....(((((....)))))(((.(..(.((((((((((...((.((((((.....))))))))))).)).))))))..).)))))).)))))))))).))). ( -52.60) >Bsubt.0 32193 120 + 4214630/2160-2280 GCGCCAGCUUCGGUGGAGGCAUCGGUGGGAUACUACCCUGGCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCC (((((.((((((((.(((.....(((((....)))))(((.(..(.((((((((((...((.((((((.....))))))))))).)).))))))..).)))))).)))))))))).))). ( -54.10) >Bcere.0 31024 120 + 5224283/2160-2280 GCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCC (((((.((....))...))))).((((....((((((((((((((.((((....((......((((((.....))))))(....)))..))))....)))))).....)))))))))))) ( -50.90) >Banth.0 31045 120 + 5227293/2160-2280 GCGCUAGCUUCGGUGGAGGCGCUGGUGGGAUACCGCCCUGACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCC (((((.((....))...))))).((((....((((((((((((((.((((....((......((((((.....))))))(....)))..))))....)))))).....)))))))))))) ( -50.90) >consensus GCGCCAGCUUCGGUGGAGGCACCGGUGGGAUACCACCCUGACUGUAUUGAAAUUCUAACCCACGGCCCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCC (((((.((((((((.((.((.(((((((....)))))(((((....................((((((.....))))))(....)...)))))..))))...)).)))))))))).))). (-50.50 = -47.38 + -3.12)


| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -47.03 |
| Consensus MFE | -45.60 |
| Energy contribution | -44.98 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.97 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916864 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2200-2320 GCUGUAUUGACCUUCUAACCCGCUGCCCUUAUCGGGCAGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAA .(((....((((((...((((.((((((.....)))))).).(((...))).)))((((.....(((((...)))))((((((...........)))))))))).))))))....))).. ( -49.10) >Bsubt.0 32193 120 + 4214630/2200-2320 GCUGUAUUGACCUUCUAACCCGCCGCCCUUAUCGGGCGGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAA .(((....((((((...((((.((((((.....)))))).).(((...))).)))((((.....(((((...)))))((((((...........)))))))))).))))))....))).. ( -50.60) >Bcere.0 31024 120 + 5224283/2200-2320 ACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAA .(((....(((......((((.((((((.....))))))(....)......))))((((.....(((((...)))))((((((...........)))))))))).....)))...))).. ( -44.20) >Banth.0 31045 120 + 5227293/2200-2320 ACUGUAUUGAAAUUCUAACCUACGGGUCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAA .(((....(((......((((.((((((.....))))))(....)......))))((((.....(((((...)))))((((((...........)))))))))).....)))...))).. ( -44.20) >consensus ACUGUAUUGAAAUUCUAACCCACGGCCCUUAUCGACCCGGGAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAA .(((....((((((...(((..((((((.....))))))(....).......)))((((.....(((((...)))))((((((...........)))))))))).))))))....))).. (-45.60 = -44.98 + -0.62)

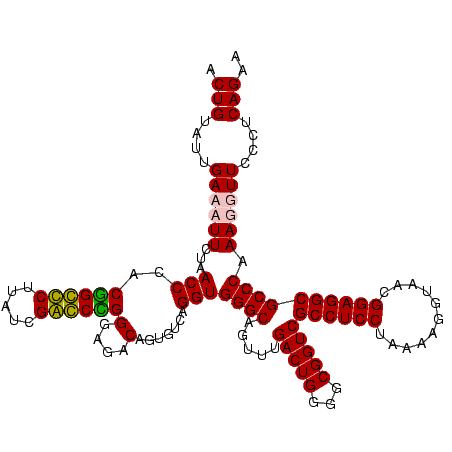
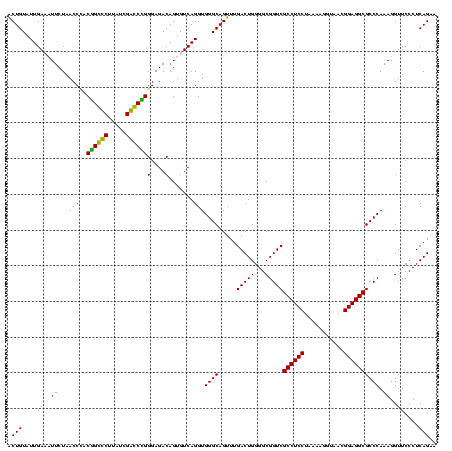
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -45.62 |
| Consensus MFE | -45.05 |
| Energy contribution | -42.93 |
| Covariance contribution | -2.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995349 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2200-2320 UUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCUGCCCGAUAAGGGCAGCGGGUUAGAAGGUCAAUACAGC ..((((..((.(((((((((((((((...........))))))(((.......))).....))))).(((((.(((.....((((((.....))))))))))))))))))))..).))). ( -47.00) >Bsubt.0 32193 120 + 4214630/2200-2320 UUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCCGCCCGAUAAGGGCGGCGGGUUAGAAGGUCAAUACAGC ..((((..((.(((((((((((((((...........))))))(((.......))).....))))).(((((.(((.....((((((.....))))))))))))))))))))..).))). ( -48.50) >Bcere.0 31024 120 + 5224283/2200-2320 UUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGU ((((((((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....))))).....))))))............ ( -43.50) >Banth.0 31045 120 + 5227293/2200-2320 UUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGGUCGAUAAGACCCGUAGGUUAGAAUUUCAAUACAGU ((((((((((.....(((((((((((...........))))))(((.......))).....)))))...(((...)))))))(((((.....))))).....))))))............ ( -43.50) >consensus UUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUCCCGGCCCGAUAAGACCCGCAGGUUAGAAGGUCAAUACAGC ..(((.(((.......((((((((((...........))))))....))))(((.....))))))..(((((.(((.....((((((.....))))))))))))))..........))). (-45.05 = -42.93 + -2.12)

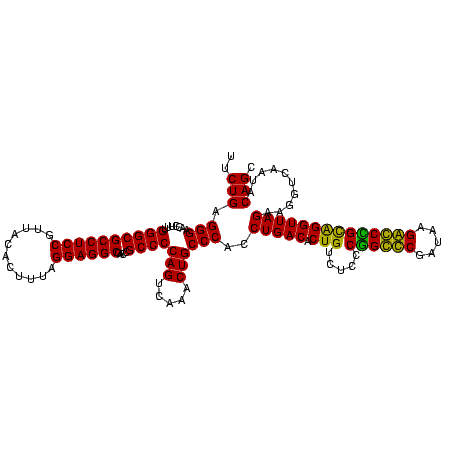
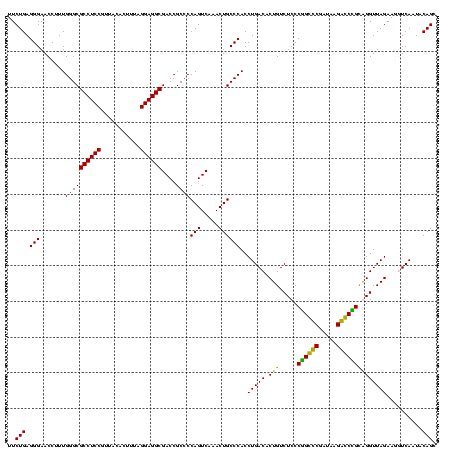
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -42.55 |
| Consensus MFE | -43.05 |
| Energy contribution | -42.55 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.96 |
| Structure conservation index | 1.01 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2240-2360 GAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAG ..(((...)))...(((((.....(((((...)))))((((((...........))))))))))).....(((((..(((((((.....))))))).....((((....)))).))))). ( -42.80) >Bsubt.0 32193 120 + 4214630/2240-2360 GAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAG ..(((...)))...(((((.....(((((...)))))((((((...........))))))))))).....(((((..(((((((.....))))))).....((((....)))).))))). ( -42.80) >Bcere.0 31024 120 + 5224283/2240-2360 GAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAG ..(((...)))...(((((.....(((((...)))))((((((...........))))))))))).....(((((..(((((((.....))))))).....((((....)))).))))). ( -42.30) >Banth.0 31045 120 + 5227293/2240-2360 GAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAG ..(((...)))...(((((.....(((((...)))))((((((...........))))))))))).....(((((..(((((((.....))))))).....((((....)))).))))). ( -42.30) >consensus GAGACAGUGUCAGGUGGGCAGUUUGACUGGGGCGGUCGCCUCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGCAAAGGCACAAGGGAG ..(((...)))...(((((.....(((((...)))))((((((...........))))))))))).....(((((..(((((((.....))))))).....((((....)))).))))). (-43.05 = -42.55 + -0.50)

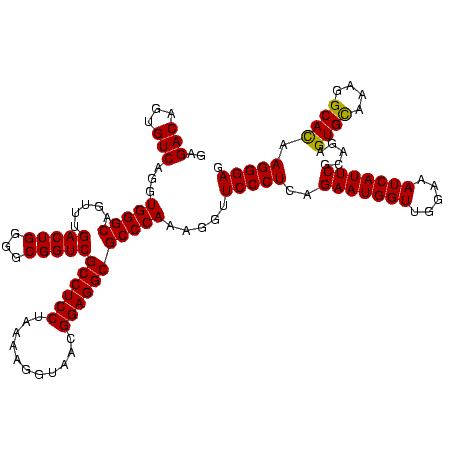
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -36.50 |
| Consensus MFE | -34.40 |
| Energy contribution | -34.90 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.94 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.974018 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2240-2360 CUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUC .(((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))).. ( -36.20) >Bsubt.0 32193 120 + 4214630/2240-2360 CUCCCUUGUGCCUUUACACUCUGCGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACCUUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUC .(((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))).. ( -36.20) >Bcere.0 31024 120 + 5224283/2240-2360 CUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUC .((((((((((....)))......(((((.........)))))))))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))).. ( -36.80) >Banth.0 31045 120 + 5227293/2240-2360 CUCCCUUAUGCCUUUGCACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUC .((((((((((....)))......(((((.........)))))))))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))).. ( -36.80) >consensus CUCCCUUAUGCCUUUACACUCUACGAAUGAUUUCCAACCAUUCUGAGGGAACCUUUGGGCGCCUCCGUUACACUUUAGGAGGCGACCGCCCCAGUCAAACUGCCCACCUGACACUGUCUC .(((((((((......))).....(((((.........))))).)))))).....(((((((((((...........))))))(((.......))).....)))))...(((...))).. (-34.40 = -34.90 + 0.50)


| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.22 |
| Mean single sequence MFE | -38.45 |
| Consensus MFE | -38.55 |
| Energy contribution | -38.05 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.89 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2280-2400 UCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUC .((.....))....((......))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))) ( -38.40) >Bsubt.0 32193 120 + 4214630/2280-2400 UCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUC .((.....))....((......))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))) ( -38.40) >Bcere.0 31024 120 + 5224283/2280-2400 UCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUC .......((((.......))))....(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))) ( -38.50) >Banth.0 31045 120 + 5227293/2280-2400 UCCUAAAGUGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUC .......((((.......))))....(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))) ( -38.50) >consensus UCCUAAAAGGUAACGGAGGCGCCCAAAGGUUCCCUCAGAAUGGUUGGAAAUCAUUCGCAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUC ..............((......))..(((((((((..(((((((.....))))))).....((((....)))).)))))))))..((((..(((......)))..)))).(((....))) (-38.55 = -38.05 + -0.50)

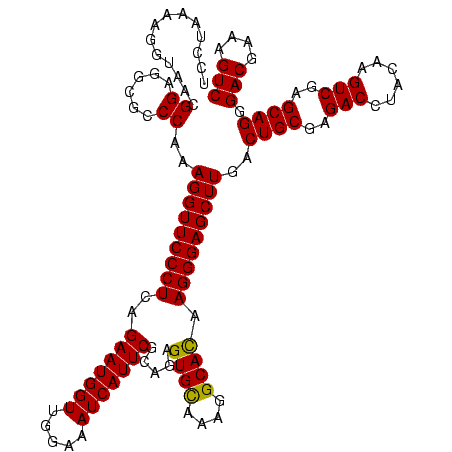
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -41.40 |
| Consensus MFE | -41.75 |
| Energy contribution | -41.25 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.54 |
| Structure conservation index | 1.01 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2320-2440 UGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCG .....(((......((((...((((....))))....((((((..((((..(((......)))..)))).(((....))))))))).)))))))((((((.((........)))))))). ( -41.80) >Bsubt.0 32193 120 + 4214630/2320-2440 UGGUUGGAAAUCAUUCGCAGAGUGUAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCG .....(((......((((...((((....))))....((((((..((((..(((......)))..)))).(((....))))))))).)))))))((((((.((........)))))))). ( -41.80) >Bcere.0 31024 120 + 5224283/2320-2440 UGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCG .....(((..(((((......((((....))))....((((((..((((..(((......)))..)))).(((....)))))))))))))))))((((((.((........)))))))). ( -41.00) >Banth.0 31045 120 + 5227293/2320-2440 UGGUUGGAAAUCAUUCGUAGAGUGCAAAGGCAUAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCG .....(((..(((((......((((....))))....((((((..((((..(((......)))..)))).(((....)))))))))))))))))((((((.((........)))))))). ( -41.00) >consensus UGGUUGGAAAUCAUUCGCAGAGUGCAAAGGCACAAGGGAGCUUGACUGCGAGACCUACAAGUCGAGCAGGGACGAAAGUCGGGCUUAGUGAUCCGGUGGUUCCGCAUGGAAGGGCCAUCG .....(((..(((((......((((....))))....((((((..((((..(((......)))..)))).(((....)))))))))))))))))((((((.((........)))))))). (-41.75 = -41.25 + -0.50)

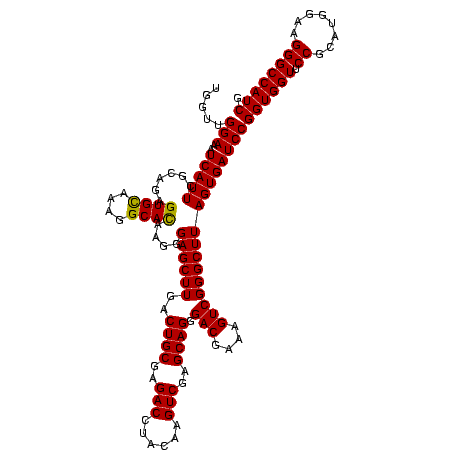
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -55.70 |
| Consensus MFE | -52.70 |
| Energy contribution | -55.70 |
| Covariance contribution | 3.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.819741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2520-2640 GAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCG ((((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))((..(.(((((((....))).)))).)..))....)))). ( -55.70) >Bsubt.0 32193 120 + 4214630/2520-2640 GAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCG ((((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))((..(.(((((((....))).)))).)..))....)))). ( -55.70) >Bcere.0 31024 120 + 5224283/2520-2640 GAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCG ((((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))((..(.(((((((....))).)))).)..))....)))). ( -55.70) >Banth.0 31045 120 + 5227293/2520-2640 GAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCG ((((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))((..(.(((((((....))).)))).)..))....)))). ( -55.70) >consensus GAUGUCGGCUCAUCGCAUCCUGGGGCUGUAGUCGGUCCCAAGGGUUGGGCUGUUCGCCCAUUAAAGCGGUACGCGAGCUGGGUUCAGAACGUCGUGAGACAGUUCGGUCCCUAUCCGUCG ((((.(((((((((((((((((((((((....))))))).)))))(((((.....))))).....)))))....))))))((..(.(((((((....))).)))).)..))....)))). (-52.70 = -55.70 + 3.00)

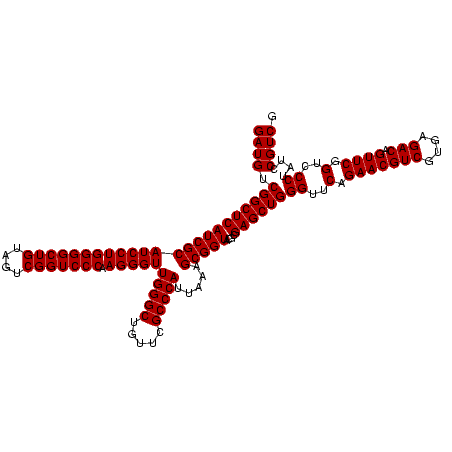
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.61 |
| Mean single sequence MFE | -31.02 |
| Consensus MFE | -32.95 |
| Energy contribution | -30.45 |
| Covariance contribution | -2.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.37 |
| Structure conservation index | 1.06 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821370 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2760-2880 UGUGUCGCCACACUUCCACCUCAGACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUAUAGGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCAC .((((....)))).(((.((((((.........(((((.......)))))..(((((.((((.....))))))))).........)))))))))(((....((......))....))).. ( -31.70) >Bsubt.0 32193 119 + 4214630/2760-2880 UGUGUCGCCACACUUCCACCUCAGACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUUCCUUGC-GGAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCAC .((((....)))).(((.((((((.........(((((.......)))))..(((((.((((....-))))))))).........)))))))))(((....((......))....))).. ( -31.40) >Bcere.0 31024 119 + 5224283/2760-2880 CAUGUCACCAUGCUUCCACCUCGAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCAC ((((....))))..(((.((((((.........(((((.......)))))..(((((.((((....-))))))))).........)))))))))(((....((......))....))).. ( -30.50) >Banth.0 31045 119 + 5227293/2760-2880 CAUGUCACCAUGCUUCCACCUCGAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUAGCUUAC-GCUAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCAC ((((....))))..(((.((((((.........(((((.......)))))..(((((.((((....-))))))))).........)))))))))(((....((......))....))).. ( -30.50) >consensus CAUGUCACCACACUUCCACCUCAAACCUAUCAACCUGAUCAUCUUUCAGGGAUCUUACUACCUUAC_GCAAUGGGAAAUCUCAUCUUGAGGGGGGCUUCAUGCUUAGAUGCUUUCAGCAC ((((....))))..(((.((((((.........(((((.......)))))..(((((.((((.....))))))))).........)))))))))(((....((......))....))).. (-32.95 = -30.45 + -2.50)

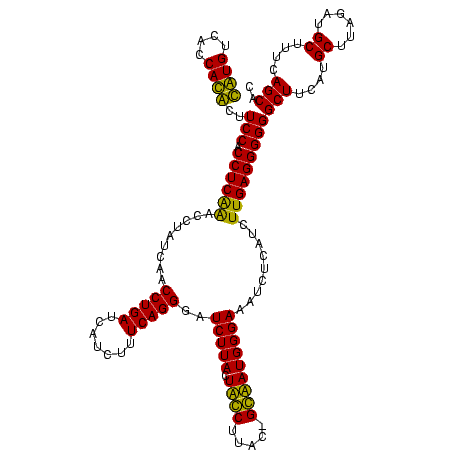
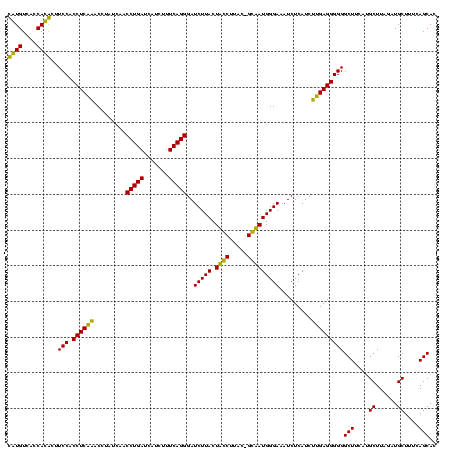
| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.36 |
| Mean single sequence MFE | -35.17 |
| Consensus MFE | -32.88 |
| Energy contribution | -31.38 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2800-2920 GAUUUCCCAUUCCUAUAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACCUAA ....((((.((((.....))))....(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))).))).......... ( -33.10) >Bsubt.0 32193 116 + 4214630/2800-2920 GAUUUCCCAUUCC-GCAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACC--- ....((((.((((-....))))....(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))).))).......--- ( -36.60) >Bcere.0 31024 118 + 5224283/2800-2920 GAUUUCCCAUAGC-GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC-AU ....((((.((((-....))))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).))).......-.. ( -35.50) >Banth.0 31045 118 + 5227293/2800-2920 GAUUUCCCAUAGC-GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC-AU ....((((.((((-....))))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).))).......-.. ( -35.50) >consensus GAUUUCCCAUACC_GUAAGCAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCCGAGGUGGAAGCAUGGCGACACAUGGAGCUGACAAAUACUAAUAGAUCGAGGACUUAACC_AU ....((((.((((.....))))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))).))).......... (-32.88 = -31.38 + -1.50)

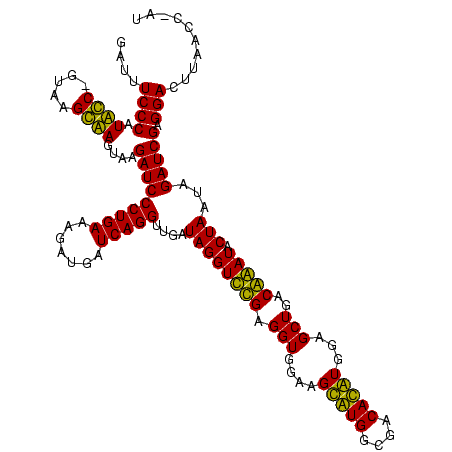

| Location | 36,508 – 36,628 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 88.38 |
| Mean single sequence MFE | -34.38 |
| Consensus MFE | -32.08 |
| Energy contribution | -30.57 |
| Covariance contribution | -1.50 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Blich.0 36508 120 + 4222334/2805-2925 CCCAUUCCUAUAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACCUAAAUAUU .((.((((.....))))....(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))..))................ ( -32.30) >Bsubt.0 32193 116 + 4214630/2805-2925 CCCAUUCC-GCAAGGAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCUGAGGUGGAAGUGUGGCGACACAUGGAGCUGACAGAUACUAAUCGAUCGAGGACUUAACC---ACAUU .((.((((-....))))....(((((((((.......)))))..((((((((((.(((....(((((....)))))...)))..))))).)).)))))))..))........---..... ( -35.80) >Bcere.0 31024 118 + 5224283/2805-2925 CCCAUAGC-GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC-AUAUAAU .((.((((-....))))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))..))........-....... ( -34.70) >Banth.0 31045 118 + 5227293/2805-2925 CCCAUAGC-GUAAGCUAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUUCGAGGUGGAAGCAUGGUGACAUGUGGAGCUGACGAAUACUAAUAGAUCGAGGACUUAACC-AUAUAAU .((.((((-....))))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))..))........-....... ( -34.70) >consensus CCCAUACC_GUAAGCAAGUAAGAUCCCUGAAAGAUGAUCAGGUUGAUAGGUCCGAGGUGGAAGCAUGGCGACACAUGGAGCUGACAAAUACUAAUAGAUCGAGGACUUAACC_AUAUAAU .((.((((.....))))....(((((((((.......)))))....((((((((.(((....(((((....)))))...)))..))))).)))...))))..))................ (-32.08 = -30.57 + -1.50)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:22:43 2006