| Sequence ID | Bcere.0 |
|---|---|
| Location | 4 – 123 |
| Length | 119 |
| Max. P | 0.999691 |

| Location | 4 – 121 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.19 |
| Mean single sequence MFE | -24.03 |
| Consensus MFE | -14.24 |
| Energy contribution | -16.05 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.77 |
| SVM RNA-class probability | 0.996901 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 4 117 + 5224283/0-120 UUUUUCUUGUUUUUUA-UAUCCACAAACUCUUUUCGUA-CUUUUACACAGUAUAUCGUGUUGUGGACAAG-UUUAUUCCACAAGGUAUUGAUUUUGUGGAUAACUUUCUUAAUUUCAUUG ................-((((((((((.((.....(((-((((.((((........)))).(((((....-.....)))))))))))).)).)))))))))).................. ( -25.10) >Banth.0 4 117 + 5227293/0-120 UUUUUCUUGUUUUUUA-UAUCCACAAACUCUUUUCGUA-CUUUUACACAGUAUAUCGUGUUGUGGACAAU-UUUAUUCCACAAGGUAUUGAUUUUGUGGAUAACUUUCUUAAUUUCAUUG ................-((((((((((.((.....(((-((((.((((........)))).(((((....-.....)))))))))))).)).)))))))))).................. ( -25.10) >Blich.0 100 119 + 4222334/0-120 UUCUCCUCGUUUUUUC-UAUCCACAAUCAUUAUCGACAACAUUUUCACAAAACUAACCGUUGUGGACAAUAUUUCUUCAACAGUUUGUAUAGUAUGUGGAUAAGUUCCUCGCAACCAUUG ................-((((((((.........((........)).....((.(((.((((.(((.......))).)))).))).))......)))))))).................. ( -17.10) >Bsubt.0 3 120 + 4214630/0-120 UUUUCGGCUUUUUUUAGUAUCCACAGAGGUUAUCGACAACAUUUUCACAUUACCAACCCCUGUGGACAAGGUUUUUUCAACAGGUUGUCCGCUUUGUGGAUAAGAUUGUGACAACCAUUG ......(((......))).(((((((.((((.......................)))).)))))))...((((...(((.(((.((((((((...))))))))..)))))).)))).... ( -28.80) >consensus UUUUCCUCGUUUUUUA_UAUCCACAAACUCUAUCCACA_CAUUUACACAGUACAAACCGUUGUGGACAAG_UUUAUUCAACAAGGUAUUGAUUUUGUGGAUAACUUUCUUAAAACCAUUG .................(((((((((((((.....(((((...................(((((((..........)))))))))))).))))))))))))).................. (-14.24 = -16.05 + 1.81)

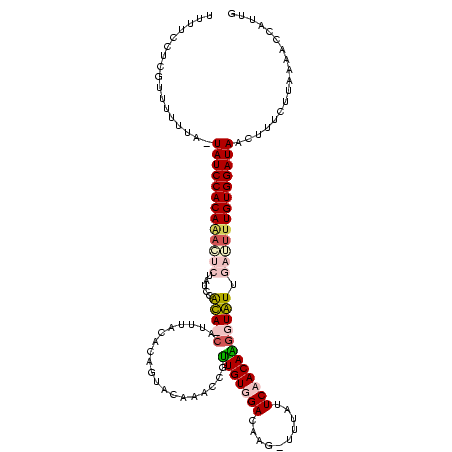

| Location | 4 – 121 |
|---|---|
| Length | 117 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 71.19 |
| Mean single sequence MFE | -27.10 |
| Consensus MFE | -12.80 |
| Energy contribution | -16.43 |
| Covariance contribution | 3.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.14 |
| Structure conservation index | 0.47 |
| SVM decision value | 3.11 |
| SVM RNA-class probability | 0.998463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 4 117 + 5224283/0-120 CAAUGAAAUUAAGAAAGUUAUCCACAAAAUCAAUACCUUGUGGAAUAAA-CUUGUCCACAACACGAUAUACUGUGUAAAAG-UACGAAAAGAGUUUGUGGAUA-UAAAAAACAAGAAAAA ..................((((((((((.((.......((((((.....-....))))))(((((......))))).....-........)).))))))))))-................ ( -24.60) >Banth.0 4 117 + 5227293/0-120 CAAUGAAAUUAAGAAAGUUAUCCACAAAAUCAAUACCUUGUGGAAUAAA-AUUGUCCACAACACGAUAUACUGUGUAAAAG-UACGAAAAGAGUUUGUGGAUA-UAAAAAACAAGAAAAA ..................((((((((((.((.......((((((.....-....))))))(((((......))))).....-........)).))))))))))-................ ( -24.60) >Blich.0 100 119 + 4222334/0-120 CAAUGGUUGCGAGGAACUUAUCCACAUACUAUACAAACUGUUGAAGAAAUAUUGUCCACAACGGUUAGUUUUGUGAAAAUGUUGUCGAUAAUGAUUGUGGAUA-GAAAAAACGAGGAGAA .................(((((((((...........((.....))...(((((((.((((((.(((......)))...)))))).)))))))..))))))))-)............... ( -22.00) >Bsubt.0 3 120 + 4214630/0-120 CAAUGGUUGUCACAAUCUUAUCCACAAAGCGGACAACCUGUUGAAAAAACCUUGUCCACAGGGGUUGGUAAUGUGAAAAUGUUGUCGAUAACCUCUGUGGAUACUAAAAAAAGCCGAAAA (((((((((((.....(((.......)))..))))))).)))).........(((((((((((((((..(((((....)))))..)...))))))))))))))................. ( -37.20) >consensus CAAUGAAAGUAAGAAACUUAUCCACAAAAUCAACAACCUGUGGAAUAAA_AUUGUCCACAACACGAUAUACUGUGAAAAAG_UACCAAAAAAGUUUGUGGAUA_UAAAAAACAAGAAAAA ..................(((((((((((((......(((((((..........)))))))((((......))))...............)))))))))))))................. (-12.80 = -16.43 + 3.62)

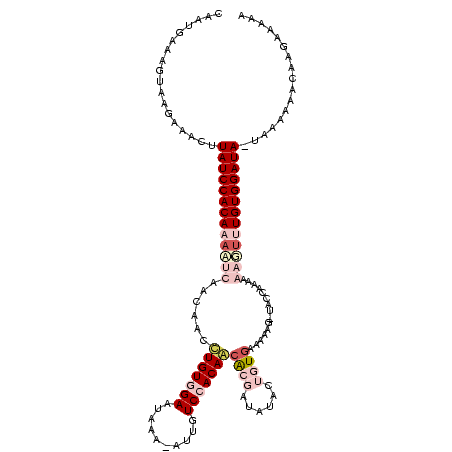
| Location | 4 – 123 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.51 |
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -10.24 |
| Energy contribution | -10.43 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 4 119 + 5224283/80-200 CAAGGUAUUGAUUUUGUGGAUAACUUUCUUAAUUUCAUUGCUAUAGCUACUUUUUUUUGAUAUUAUAGUUGUGUUUUCACUUUGAAUAAGUU-UUCCACAUCUUUAUCUUAUCCACAAUU .((((((..((...((((((.(((((..((((...(((.(((((((.(((........).))))))))).)))........))))..)))))-.))))))))..)))))).......... ( -23.90) >Banth.0 4 119 + 5227293/80-200 CAAGGUAUUGAUUUUGUGGAUAACUUUCUUAAUUUCAUUGCUAUAGCUACUUUUUUUUGAUAUUAUAGUUGUGUUUUCACUUUGAAUAAGUU-UUCCACAUCUUUAUCUUAUCCACAAUU .((((((..((...((((((.(((((..((((...(((.(((((((.(((........).))))))))).)))........))))..)))))-.))))))))..)))))).......... ( -23.90) >Blich.0 100 120 + 4222334/80-200 CAGUUUGUAUAGUAUGUGGAUAAGUUCCUCGCAACCAUUGCAACCACUCGCUUAUUCUGAUAUUAUAUUUGUGUUUUAACUCUUGAUAACAAAUUGGCUGCCAAUCCAUUAUCCACAAAC ...........((.(((((((((.......(((.(((..((........))...............((((((...(((.....)))..))))))))).))).......))))))))).)) ( -21.74) >Bsubt.0 3 120 + 4214630/80-200 CAGGUUGUCCGCUUUGUGGAUAAGAUUGUGACAACCAUUGCAAGCUCUCGUUUAUUUUGGUAUUAUAUUUGUGUUUUAACUCUUGAUUACUAAUCCUACCUUUCCUCUUUAUCCACAAAG ..((....)).((((((((((((((((((((((((((..(.((((....)))).)..)))).........((......))...)).)))).)))).............)))))))))))) ( -23.31) >consensus CAAGGUAUUGAUUUUGUGGAUAACUUUCUUAAAACCAUUGCAAUAGCUACUUUAUUUUGAUAUUAUAGUUGUGUUUUAACUCUGAAUAACUA_UUCCACAUCUUUACCUUAUCCACAAAU ...........((((((((((((...................................(((((.......)))))(((.....)))......................)))))))))))) (-10.24 = -10.43 + 0.19)

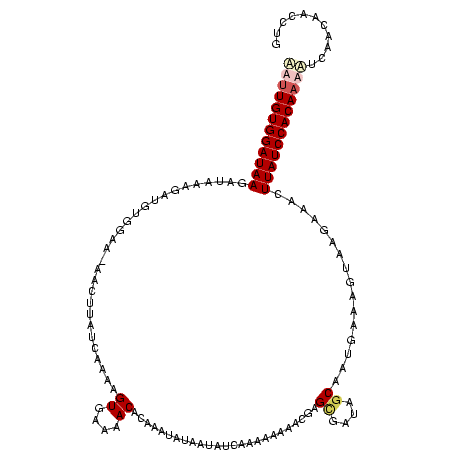
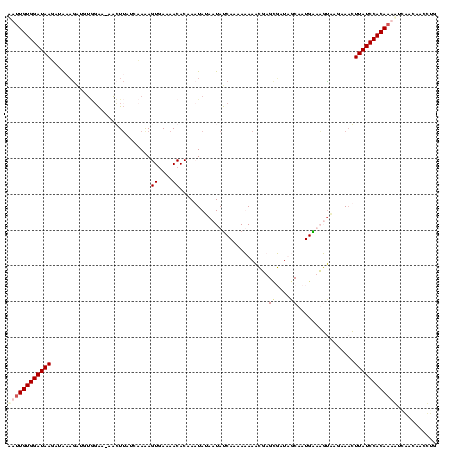
| Location | 4 – 123 |
|---|---|
| Length | 119 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.51 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -11.82 |
| Energy contribution | -13.08 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.96 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 4 119 + 5224283/80-200 AAUUGUGGAUAAGAUAAAGAUGUGGAA-AACUUAUUCAAAGUGAAAACACAACUAUAAUAUCAAAAAAAAGUAGCUAUAGCAAUGAAAUUAAGAAAGUUAUCCACAAAAUCAAUACCUUG ..((((((((((((((.((.((((...-.((((.....)))).....)))).))....))))...........((....))................))))))))))............. ( -22.30) >Banth.0 4 119 + 5227293/80-200 AAUUGUGGAUAAGAUAAAGAUGUGGAA-AACUUAUUCAAAGUGAAAACACAACUAUAAUAUCAAAAAAAAGUAGCUAUAGCAAUGAAAUUAAGAAAGUUAUCCACAAAAUCAAUACCUUG ..((((((((((((((.((.((((...-.((((.....)))).....)))).))....))))...........((....))................))))))))))............. ( -22.30) >Blich.0 100 120 + 4222334/80-200 GUUUGUGGAUAAUGGAUUGGCAGCCAAUUUGUUAUCAAGAGUUAAAACACAAAUAUAAUAUCAGAAUAAGCGAGUGGUUGCAAUGGUUGCGAGGAACUUAUCCACAUACUAUACAAACUG ((.(((((((((.(..((.(((((((..((((.((((...(((.........................)))...)))).))))))))))).))...)))))))))).))........... ( -31.01) >Bsubt.0 3 120 + 4214630/80-200 CUUUGUGGAUAAAGAGGAAAGGUAGGAUUAGUAAUCAAGAGUUAAAACACAAAUAUAAUACCAAAAUAAACGAGAGCUUGCAAUGGUUGUCACAAUCUUAUCCACAAAGCGGACAACCUG ((((((((((((.((.....((((.(((.....)))....((....))..........)))).........((.((((......)))).))....)))))))))))))).((....)).. ( -25.50) >consensus AAUUGUGGAUAAGAUAAAGAUGUGGAA_AACUUAUCAAAAGUGAAAACACAAAUAUAAUAUCAAAAAAAACGAGCGAUAGCAAUGAAAGUAAGAAACUUAUCCACAAAAUCAACAACCUG ((((((((((((............................((....)).........................((....))................))))))))))))........... (-11.82 = -13.08 + 1.25)

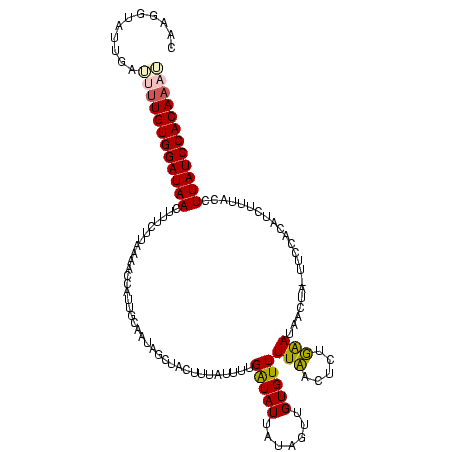
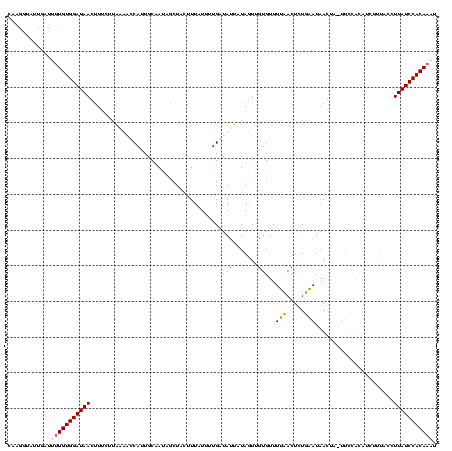
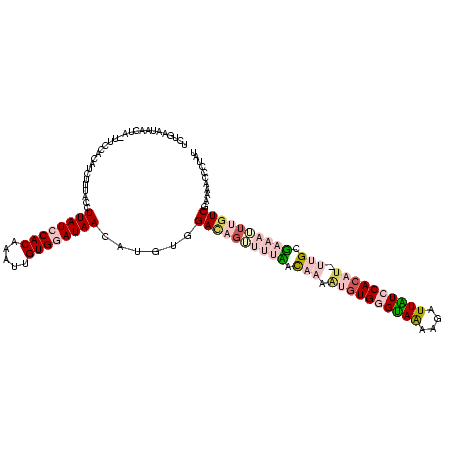
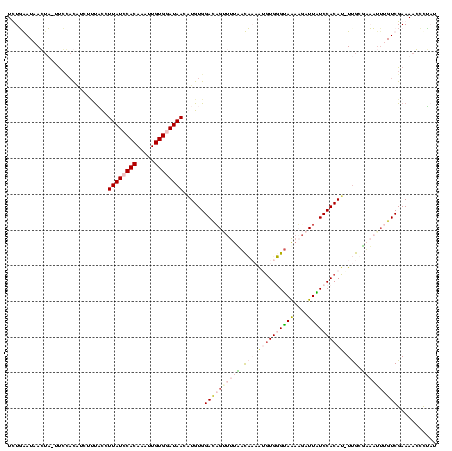
| Location | 4 – 122 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 66.57 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -15.68 |
| Energy contribution | -19.80 |
| Covariance contribution | 4.12 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.51 |
| Structure conservation index | 0.49 |
| SVM decision value | 3.90 |
| SVM RNA-class probability | 0.999691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 4 118 + 5224283/160-280 UUUGAAUAAGUU-UUCCACAUCUUUAUCUUAUCCACAAUUUGUGUAUAACAUGUGGACAGUUUUAAUCACAUGUGGGUAAAUGAUUAUCCACAU-UCGCUUUUUUUGUCGAAAACCCUAU .........(((-(((.(((...........((((((...(((.....))))))))).............((((((((((....))))))))))-..........))).))))))..... ( -26.00) >Banth.0 4 117 + 5227293/160-280 UUUGAAUAAGUU-UUCCACAUCUUUAUCUUAUCCACAAUUUGUGUAUAACAUGUGGACAGUUUUAAUCACAUGUGGGUAAAUGAUUAUCCACAU-UUGCUUUUUU-GUCGAAAACCCUAU .........(((-(((.(((...........((((((...(((.....)))))))))..........((.((((((((((....))))))))))-.))......)-)).))))))..... ( -27.00) >Blich.0 100 120 + 4222334/160-280 UCUUGAUAACAAAUUGGCUGCCAAUCCAUUAUCCACAAACUGUGGAUAAGUUGUGGAGAGUUUUUUCACAGGGUGUGCAGUAUUUUGUCCACAUCUUGUGAAAAAUGUCGAAAAGACGUU ((((.(((((..(((((...)))))...((((((((.....))))))))))))).))))...((((((((((((((((((....)))..)))))))))))))))(((((.....))))). ( -44.30) >Bsubt.0 3 120 + 4214630/160-280 UCUUGAUUACUAAUCCUACCUUUCCUCUUUAUCCACAAAGUGUGGAUAAGUUGUGGAUUGAUUUCACACAGCUUGUGUAGAAGGUUGUCCACAAGUUGUGAAAUUUGUCGAAAAGCUAUU ..(((((....(((..(((........(((((((((.....)))))))))((((((((...((((((((.....)))).))))...))))))))...)))..))).)))))......... ( -31.30) >consensus UCUGAAUAACUA_UUCCACAUCUUUACCUUAUCCACAAAUUGUGGAUAACAUGUGGACAGUUUUAACAAAAUGUGGGUAAAAGAUUAUCCACAU_UUGCGAAAUUUGUCGAAAACCCUAU ............................((((((((.....))))))))......((((((((((.(((.((((((((((....)))))))))).))).))))))))))........... (-15.68 = -19.80 + 4.12)

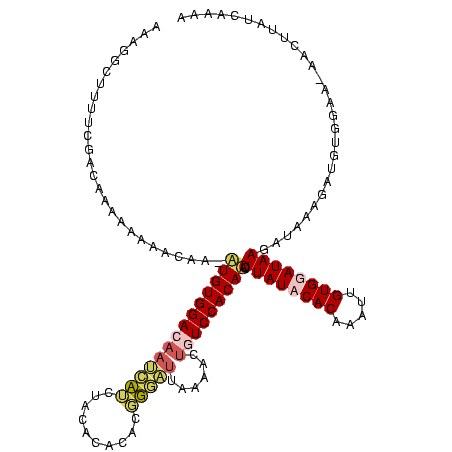
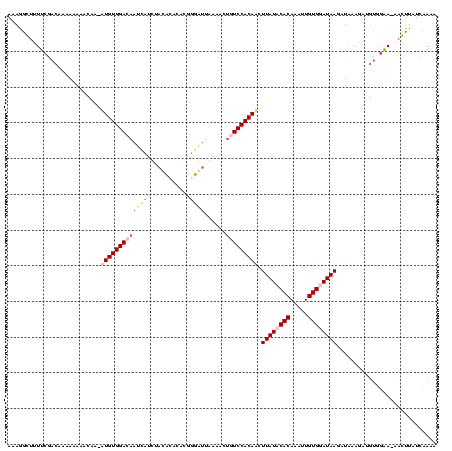
| Location | 4 – 122 |
|---|---|
| Length | 118 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 66.57 |
| Mean single sequence MFE | -30.78 |
| Consensus MFE | -13.32 |
| Energy contribution | -14.32 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -3.70 |
| Structure conservation index | 0.43 |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.998344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Bcere.0 4 118 + 5224283/160-280 AUAGGGUUUUCGACAAAAAAAGCGA-AUGUGGAUAAUCAUUUACCCACAUGUGAUUAAAACUGUCCACAUGUUAUACACAAAUUGUGGAUAAGAUAAAGAUGUGGAA-AACUUAUUCAAA ...((((((((.(((.......((.-((((((.(((....))).)))))).))........(((((((((((.....)))...)))))))).........))).)))-)))))....... ( -29.60) >Banth.0 4 117 + 5227293/160-280 AUAGGGUUUUCGAC-AAAAAAGCAA-AUGUGGAUAAUCAUUUACCCACAUGUGAUUAAAACUGUCCACAUGUUAUACACAAAUUGUGGAUAAGAUAAAGAUGUGGAA-AACUUAUUCAAA ...((((((((.((-(......((.-((((((.(((....))).)))))).))........(((((((((((.....)))...)))))))).........))).)))-)))))....... ( -30.30) >Blich.0 100 120 + 4222334/160-280 AACGUCUUUUCGACAUUUUUCACAAGAUGUGGACAAAAUACUGCACACCCUGUGAAAAAACUCUCCACAACUUAUCCACAGUUUGUGGAUAAUGGAUUGGCAGCCAAUUUGUUAUCAAGA ...(((.....))).(((((((((.(.((((.(........).)))).).)))))))))......(((((((.......)).)))))(((((..((((((...))))))..))))).... ( -31.70) >Bsubt.0 3 120 + 4214630/160-280 AAUAGCUUUUCGACAAAUUUCACAACUUGUGGACAACCUUCUACACAAGCUGUGUGAAAUCAAUCCACAACUUAUCCACACUUUGUGGAUAAAGAGGAAAGGUAGGAUUAGUAAUCAAGA ....(((((((((...((((((((.((((((............))))))...))))))))...))......((((((((.....))))))))....)))))))................. ( -31.50) >consensus AAAGGCUUUUCGACAAAAAAAACAA_AUGUGGACAAUCAUCUACACACACGGGAUUAAAACUGUCCACAACUUAUACACAAAUUGUGGAUAAGAUAAAGAUGUGGAA_AACUUAUCAAAA ..........................((((((((((((((..........)))))......))))))))).((((((((.....))))))))............................ (-13.32 = -14.32 + 1.00)

Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:22:11 2006