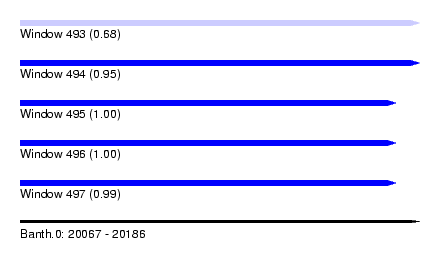
| Sequence ID | Banth.0 |
|---|---|
| Location | 20,067 – 20,186 |
| Length | 119 |
| Max. P | 0.997437 |

| Location | 20,067 – 20,186 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.58 |
| Mean single sequence MFE | -20.74 |
| Consensus MFE | -13.68 |
| Energy contribution | -9.44 |
| Covariance contribution | -4.24 |
| Combinations/Pair | 1.91 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.683660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 20067 119 + 5227293/0-120 UCAUAAAGAUAUUCCACAUU-CCAUUCCAGGAUGGAUUCACAAACUGUUUCUAUCGAUUUCCACCAACCAUCGACUCUCUAUAAGAAACGACAUUUGUUACUACUUCCUAUCAACACGCU .......((((........(-(((((....))))))...(((((.(((((((.(((((...........))))).........)))))))...)))))..........))))........ ( -15.60) >Bcere.0 20069 119 + 5224283/0-120 UCAUAAAGAUAUUCCACAUU-CCAUUCCAGGAUGGAUUCACAAACUGUUUCUAUCGAUUUCCACCAACCAUCGACUCUCUAUAAGAAACGACAUUUGUUACUAUUUCCUAUCAACACGCU .......((((........(-(((((....))))))...(((((.(((((((.(((((...........))))).........)))))))...)))))..........))))........ ( -15.60) >Bhalo.0 34410 110 + 4202352/0-120 ---------UGUUCAGCAUU-CCACUCAAGGAUGGAUUCAUAGGCACCUCUGGCUAGUUCGCACCGUCCACUAGCUCUCUGAACAGAAGUAUUCCUAUUACUAGUUCCUCUCAACGCAUU ---------(((((((....-...((...((((((...((.((....)).))((......)).))))))...))....)))))))..((((.......)))).................. ( -19.80) >Bsubt.0 1110217 115 - 4214630/0-120 C-----GGCUUUUCACCGCUGCAACUCUGAGGUGAGUUCGGAAACAAACUUGAUUGACUUUCACCAGCCGUCAACUCUCUGCAACAAUAUCUGUUUCUUACUGCUCCUCUUCAUUGUUCU (-----(((........)))).(((..(((((.((((..(((((((....(((.......)))...((.(........).)).........)))))))....))))..)))))..))).. ( -21.60) >Blich.0 1161739 115 - 4222334/0-120 C-----AGCUUAUCACUGCUGCAACUCUGAGGCGAGUUCGGAAGCUCAUUUGAUUGACUUUCACCAGCCGUCAACUCUCUGAAACAAAAUGGGCUUCUUACUGCUCCUCUUCACUGUUCU (-----(((........)))).(((..(((((.((((..(((((((((((((((((((...........))))).)).........))))))))))))....))))..)))))..))).. ( -31.10) >consensus C_____AGAUAUUCAACAUU_CCACUCCAGGAUGGAUUCACAAACUCAUUUGAUUGAUUUCCACCAACCAUCAACUCUCUGAAACAAAAUACAUUUCUUACUACUUCCUAUCAACGCGCU ............................((((.(.....(((((.(((((((.(((((...........))))).........)))))))...))))).....).))))........... (-13.68 = -9.44 + -4.24)


| Location | 20,067 – 20,186 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 58.23 |
| Mean single sequence MFE | -39.66 |
| Consensus MFE | -14.80 |
| Energy contribution | -14.20 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.78 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.37 |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.946015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 20067 119 + 5227293/80-200 UGAAUCCAUCCUGGAAUGG-AAUGUGGAAUAUCUUUAUGAUUAGUAAACAUUCCCGGUGAAGAGCCGUUAUUUCUACUUGAGAGGAAGGCGGUAAUGCUUUCAACUAGGGUGGCAACGCG .....(((((((((...((-(((((..(..(((.....)))...)..)))))))((((.....))))...((((.....)))).(((((((....)))))))..)))))))))....... ( -37.00) >Bcere.0 20069 119 + 5224283/80-200 UGAAUCCAUCCUGGAAUGG-AAUGUGGAAUAUCUUUAUGAUUAGUAAACAUUCCCGGUGAAGAGCCGUUAUUUCUACUUGAGAGGAAAGCGGUAAUGCUUUCAACUAGGGUGGCAACGCG .....(((((((((...((-(((((..(..(((.....)))...)..)))))))((((.....))))...((((.....)))).(((((((....)))))))..)))))))))....... ( -37.30) >Bhalo.0 34410 107 + 4202352/80-200 UGAAUCCAUCCUUGAGUGG-AAUGCUGAACA---------UAAGUAGGCGUUUCCGGUUCUG-GCCGUUAU--CCAGAUGAGUGGUCAGCGGUAUAGCUGGCAACAAGGGUGGCAACGCG .....(((((((((...((-((((((..((.---------...)).))))))))(((((...-((((((..--(((......)))..))))))..)))))....)))))))))....... ( -38.60) >Bsubt.0 1110217 105 - 4214630/80-200 CGAACUCACCUCAGAGUUGCAGCGGUGAAAAGCC-----GCUGACGCAUAACUGCGUUAAAA-----GUAU---CAAGUGAGUG--CGGCGUAUGCCGCGCUAACGAGGGUGGUACCGCG ........((((...(.(((((((((.....)))-----)))((((((....))))))....-----))).---)..((.((((--((((....)))))))).))))))(((....))). ( -43.90) >Blich.0 1161739 104 - 4222334/80-200 CGAACUCGCCUCAGAGUUGCAGCAGUGAUAAGCU-----GCUGCCGCGUAUCUGCGUUACAG-----AUAU---CAAGUGAGUG--UGCCGCGUGC-ACACUAACAAGGGUGGUACCGCG ..(((((......)))))((((((((.....)))-----)))))((((((((((.....)))-----))))---...((.((((--(((.....))-))))).))..((......))))) ( -41.50) >consensus UGAAUCCAUCCUAGAGUGG_AAUGGUGAAAAGCU_____AUUAGUAAACAUUUCCGGUAAAG_GCCGUUAU__CCAAGUGAGUGG_AGGCGGUAACGCUGCCAACAAGGGUGGCAACGCG .....(((((((((.((....)).............................................................(((((((....)))))))..)))))))))....... (-14.80 = -14.20 + -0.60)

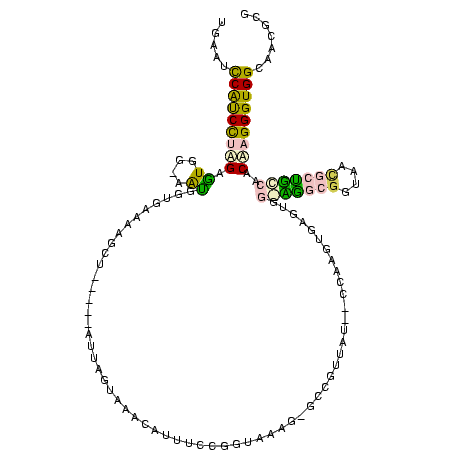
| Location | 20,067 – 20,179 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.58 |
| Mean single sequence MFE | -43.50 |
| Consensus MFE | -19.46 |
| Energy contribution | -20.54 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.50 |
| Mean z-score | -3.13 |
| Structure conservation index | 0.45 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.997009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 20067 112 + 5227293/120-240 UUAGUAAACAUUCCCGGUGAAGAGCCGUUAUUUCUACUUGAGAGGAAGGCGGUAAUGCUUUCAACUAGGGUGGCAACGCGGGUUAA-------CUCCCGUCCCUUUA-UAUAGGGACGGG ............((((((.....)))(((....(((((..((..(((((((....)))))))..))..))))).)))..)))....-------..(((((((((...-...))))))))) ( -41.50) >Bcere.0 20069 112 + 5224283/120-240 UUAGUAAACAUUCCCGGUGAAGAGCCGUUAUUUCUACUUGAGAGGAAAGCGGUAAUGCUUUCAACUAGGGUGGCAACGCGGGUUAA-------CUCCCGUCCCUUUA-UAUAGGGACGGG ............((((((.....)))(((....(((((..((..(((((((....)))))))..))..))))).)))..)))....-------..(((((((((...-...))))))))) ( -41.80) >Bhalo.0 34410 108 + 4202352/120-240 UAAGUAGGCGUUUCCGGUUCUG-GCCGUUAU--CCAGAUGAGUGGUCAGCGGUAUAGCUGGCAACAAGGGUGGCAACGCGGGU--G-------CUCUCGUCCCUUUUCUUUAGGGAUGAG ......((((((.((((((...-((((((..--(((......)))..))))))..)))))).)))....(((....)))....--)-------))(((((((((.......))))))))) ( -42.50) >Bsubt.0 1110217 108 - 4214630/120-240 CUGACGCAUAACUGCGUUAAAA-----GUAU---CAAGUGAGUG--CGGCGUAUGCCGCGCUAACGAGGGUGGUACCGCGGGAAAACGAAAGUCUCUCGUCCCUUUU-U-GGGAUGAGGG .(((((((....)))))))...-----((((---((.((.((((--((((....)))))))).)).....)))))).........((....))((((((((((....-.-)))))))))) ( -47.10) >Blich.0 1161739 101 - 4222334/120-240 CUGCCGCGUAUCUGCGUUACAG-----AUAU---CAAGUGAGUG--UGCCGCGUGC-ACACUAACAAGGGUGGUACCGCGGGAAA-------UCUCUCGUCCCUUCU-UAGGGAUGGGAG ...(((((((((((.....)))-----))))---...((.((((--(((.....))-))))).))..((......))))))....-------.(((((((((((...-.))))))))))) ( -44.60) >consensus UUAGUAAACAUUUCCGGUAAAG_GCCGUUAU__CCAAGUGAGUGG_AGGCGGUAACGCUGCCAACAAGGGUGGCAACGCGGGUAAA_______CUCUCGUCCCUUUU_UAUAGGGACGGG ............(((..........................((.(((((((....))))))).))....(((....)))))).............(((((((((.......))))))))) (-19.46 = -20.54 + 1.08)

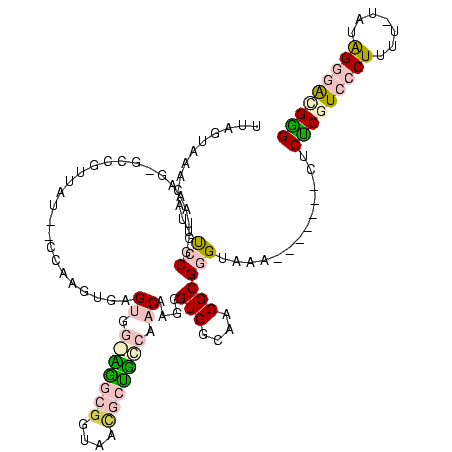
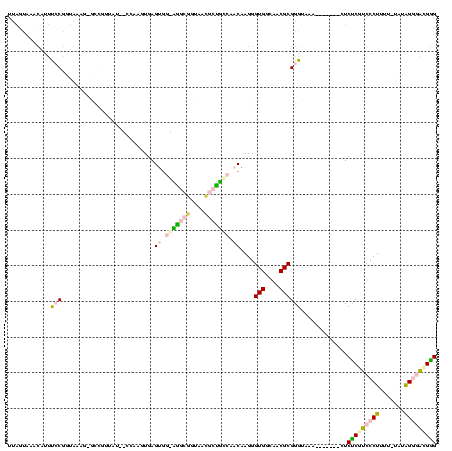
| Location | 20,067 – 20,179 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 64.53 |
| Mean single sequence MFE | -47.80 |
| Consensus MFE | -24.46 |
| Energy contribution | -25.54 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.46 |
| Mean z-score | -4.79 |
| Structure conservation index | 0.51 |
| SVM decision value | 2.86 |
| SVM RNA-class probability | 0.997437 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 20067 112 + 5227293/128-248 CAUUCCCGGUGAAGAGCCGUUAUUUCUACUUGAGAGGAAGGCGGUAAUGCUUUCAACUAGGGUGGCAACGCGGGUUAA-------CUCCCGUCCCUUUA-UAUAGGGACGGGAGUUUUUU ....((((((.....)))(((....(((((..((..(((((((....)))))))..))..))))).)))..)))..((-------(((((((((((...-...))))))))))))).... ( -48.80) >Bcere.0 20069 112 + 5224283/128-248 CAUUCCCGGUGAAGAGCCGUUAUUUCUACUUGAGAGGAAAGCGGUAAUGCUUUCAACUAGGGUGGCAACGCGGGUUAA-------CUCCCGUCCCUUUA-UAUAGGGACGGGAGUUUUUU ....((((((.....)))(((....(((((..((..(((((((....)))))))..))..))))).)))..)))..((-------(((((((((((...-...))))))))))))).... ( -49.10) >Bhalo.0 34410 108 + 4202352/128-248 CGUUUCCGGUUCUG-GCCGUUAU--CCAGAUGAGUGGUCAGCGGUAUAGCUGGCAACAAGGGUGGCAACGCGGGU--G-------CUCUCGUCCCUUUUCUUUAGGGAUGAGAGCUUUUU .(((.((((((...-((((((..--(((......)))..))))))..)))))).)))....(((....)))....--(-------(((((((((((.......))))))))))))..... ( -49.00) >Bsubt.0 1110217 108 - 4214630/128-248 UAACUGCGUUAAAA-----GUAU---CAAGUGAGUG--CGGCGUAUGCCGCGCUAACGAGGGUGGUACCGCGGGAAAACGAAAGUCUCUCGUCCCUUUU-U-GGGAUGAGGGAGUUUUUU ...(((((......-----((((---((.((.((((--((((....)))))))).)).....)))))))))))......((((.(((((((((((....-.-))))))))))).)))).. ( -49.30) >Blich.0 1161739 101 - 4222334/128-248 UAUCUGCGUUACAG-----AUAU---CAAGUGAGUG--UGCCGCGUGC-ACACUAACAAGGGUGGUACCGCGGGAAA-------UCUCUCGUCCCUUCU-UAGGGAUGGGAGAGUUUUUU ..((((((.(((..-----.(((---(..((.((((--(((.....))-))))).))...))))))).))))))...-------((((((((((((...-.))))))))))))....... ( -42.80) >consensus CAUUUCCGGUAAAG_GCCGUUAU__CCAAGUGAGUGG_AGGCGGUAACGCUGCCAACAAGGGUGGCAACGCGGGUAAA_______CUCUCGUCCCUUUU_UAUAGGGACGGGAGUUUUUU ....(((..........................((.(((((((....))))))).))....(((....))))))...........(((((((((((.......)))))))))))...... (-24.46 = -25.54 + 1.08)

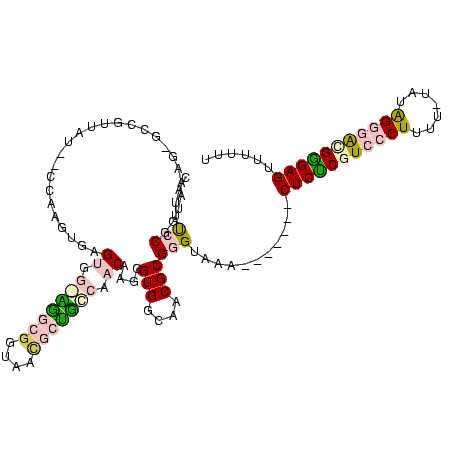
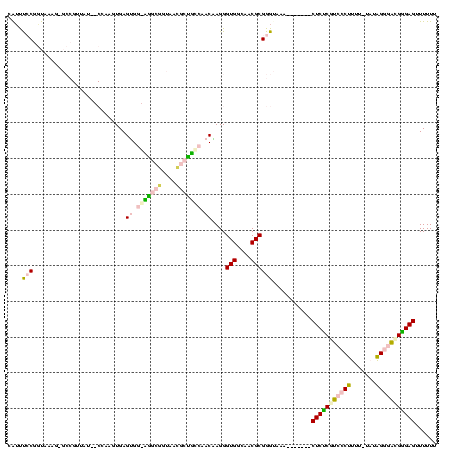
| Location | 20,067 – 20,179 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 64.53 |
| Mean single sequence MFE | -36.12 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.96 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.43 |
| Mean z-score | -3.74 |
| Structure conservation index | 0.29 |
| SVM decision value | 2.46 |
| SVM RNA-class probability | 0.994274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>Banth.0 20067 112 + 5227293/128-248 AAAAAACUCCCGUCCCUAUA-UAAAGGGACGGGAG-------UUAACCCGCGUUGCCACCCUAGUUGAAAGCAUUACCGCCUUCCUCUCAAGUAGAAAUAACGGCUCUUCACCGGGAAUG ....(((((((((((((...-...)))))))))))-------))..((((.((.(((.....((..(((.((......)).)))..))....((....))..))).....)))))).... ( -40.60) >Bcere.0 20069 112 + 5224283/128-248 AAAAAACUCCCGUCCCUAUA-UAAAGGGACGGGAG-------UUAACCCGCGUUGCCACCCUAGUUGAAAGCAUUACCGCUUUCCUCUCAAGUAGAAAUAACGGCUCUUCACCGGGAAUG ....(((((((((((((...-...)))))))))))-------))..((((.((.(((.....((..((((((......))))))..))....((....))..))).....)))))).... ( -44.70) >Bhalo.0 34410 108 + 4202352/128-248 AAAAAGCUCUCAUCCCUAAAGAAAAGGGACGAGAG-------C--ACCCGCGUUGCCACCCUUGUUGCCAGCUAUACCGCUGACCACUCAUCUGG--AUAACGGC-CAGAACCGGAAACG .....((((((.(((((.......))))).)))))-------)--..(((.((((((...........((((......)))).(((......)))--.....)))-...))))))..... ( -35.80) >Bsubt.0 1110217 108 - 4214630/128-248 AAAAAACUCCCUCAUCCC-A-AAAAGGGACGAGAGACUUUCGUUUUCCCGCGGUACCACCCUCGUUAGCGCGGCAUACGCCG--CACUCACUUG---AUAC-----UUUUAACGCAGUUA ....(((((.(((.((((-.-....)))).))).)).............(((...........((.((.(((((....))))--).)).))(((---(...-----..))))))).))). ( -27.60) >Blich.0 1161739 101 - 4222334/128-248 AAAAAACUCUCCCAUCCCUA-AGAAGGGACGAGAGA-------UUUCCCGCGGUACCACCCUUGUUAGUGU-GCACGCGGCA--CACUCACUUG---AUAU-----CUGUAACGCAGAUA ......(((((...(((((.-...))))).))))).-------....(((((((((.((........))))-)).)))))..--..........---.(((-----(((.....)))))) ( -31.90) >consensus AAAAAACUCCCGUCCCUAUA_AAAAGGGACGAGAG_______UUAACCCGCGUUGCCACCCUAGUUGAAAGCACUACCGCCG_CCACUCAAGUGG__AUAACGGC_CUUCACCGGAAAUG ......(((((.(((((.......))))).))))).................................((((......))))...................................... (-10.52 = -10.96 + 0.44)


Generated by rnazCluster.pl (part of RNAz $RNAz::rnazVersion) on Thu Apr 6 17:22:04 2006